
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Tzu-Hua Chen-Liang | + 1130 word(s) | 1130 | 2021-05-24 10:58:08 | | | |
| 2 | Lindsay Dong | Meta information modification | 1130 | 2021-05-24 11:40:44 | | |
Video Upload Options
Myelodysplastic syndromes (MDS) are a group of clonal hematopoietic neoplasms characterized by ineffective hematopoiesis and myelodysplasia with a variable spectrum of clinical–biological features that can be used to build a prognostic estimation.
1. Introduction
Since the publication of the International Prognosis Scoring System (IPSS) in 1997 [1], the international community has focused on finding the best scheme to delineate patient prognosis and cover all subcategories of this group of neoplasms. Nevertheless, with the advent of new massive sequencing tools and their constant improvement, we have a collection of genetic information in terms of the presence of mutations that may change former prognostic scores. Prognostic tools, although designed for most medical conditions, perform better in entities with a heterogeneous clinical course. Myelodysplastic syndromes (MDS) are arguably the most heterogeneous entity among hematological neoplasms. Every day in hematologist waiting rooms, we find patients with the same diagnostic label but very different plans of treatment, from wait-and-watch visits every six months with just an automatized blood count to patients going through the extremely complex road of an allogenic hematopoietic transplantation.
In daily clinical practice, current prognostic scoring systems are built up from two types of factors: patient (or host) and MDS-related [2]. With regard to MDS-related factors, peripheral blood and bone marrow features constitute the main source of predictors. In this category, cytogenetics stands as the most relevant factor in IPSS-R (Figure 1).
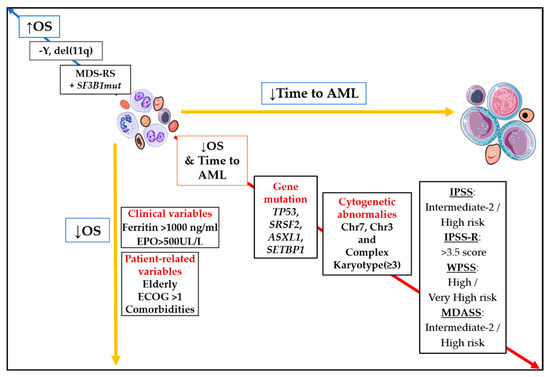
Figure 1. Impact of main prognostic markers on overall survival (OS) and time to acute myeloid leukemia (AML). Diagonal red arrow indicates factors associated both with worse OS and shorter time to AML. Opposite blue arrow is pointing out main factors are associated with better survival. Abbreviations: AML, acute myeloid leukemia; OS, overall survival; Chr, chromosome; EPO, erythropoietin; ECOG, Eastern Cooperative Oncology Group; MDS-RS, myelodysplastic syndrome with ring sideroblasts; −Y, monosomy Y; del(11q), deletion of long arm of chromosome 11; ↓, inferior; ↑; superior.
2. Incoming MDS Prognostic Models
Next-generation sequencing has allowed for large-scale analysis of the molecular profile in MDS. Not surprisingly, its mutational landscape is quite heterogeneous, something that is considered critical to define MDS clinical and pathological features since its first description. The collection of NGS data has granted significant knowledge about the pathophysiological complexity of this blood cancer, involving a set of recurrently mutated genes that play diverse roles in cellular processes, such as DNA methylation, chromatin modification, signal transduction, transcriptional regulation, and RNA splicing [3][4][5][6][7]. Among patients diagnosed with MDS, more than 50% have normal cytogenetics, the only genomic variable included in current prognostic scores, but about 90% harbor mutations. The mutational burden is directly correlated with the number of karyotypic aberrations. Still, a high percentage of low-risk and normal karyotype cases present with an average of 1–3 somatic mutations [8][9].
From a strict quantitative view, the more acquired variants, the more likely the MDS to behave aggressively. Both Bejar and Papaemmanuil showed in a large series of MDS cases that stratification by the number of tumor mutations was highly predictive of LFS. Harboring three or more acquired mutations defined a subset of cases with approximately two years to leukemia transformation [10][11].
Another quantitative parameter, the variant allele frequency (VAF), has been studied for prognostic purposes. VAF depicts the percentage of mutated cells within the sample, and it is used as a surrogate marker to define clonal (predominant) or subclonal tumor population. However, for the VAF to be considered as a prognostic factor, the gene affected must be taken into account. TP53, TET2 and SF3B1 prognostic impact has been shown to be modified by the VAF. On the other hand, there are other targets, i.e., NRAS and EZH2, that seems to predict for more aggressive course regardless of the VAF [12][13][14][15].
Seven genes were previously reported to harbor independent prognostic significance in MDS and impact on OS: TP53, EZH2, RUNX1, NRAS, ASXL1 and SF3B1. All of them, regardless of SF3B1, have negative influence on survival and could actively participate as triggers of disease progression to AML [10][16][17]. However, not a single or group of gene mutations were included in the IPSS-R (Figure 1).
Using more refined statistical approaches, a recent study addressed the likely underestimated importance of mutation co-occurrence patterns in MDS prognostic settings [18]. Though external validation is needed, the authors stated that specific comutation patterns account for clinical heterogeneity within SF3B1, SRSF2, and del5q MDS. In addition, acquired mutations may be useful for both anticipating the natural history of the disease at diagnosis, and minimal residual disease assessment and prediction of progression during the course of MDS [19][20][21]. Lastly, the success or failure of emerging targeted therapies (i.e., IDH1/2 or spliceosome inhibitors) may accelerate the implementation of new genes in the workup on MDS patients. Huge effort is currently expended to reach a molecular IPSS-R.
Regarding the allele state of TP53, a recent study reported robust association between TP53 biallelic/multihit mutations and an inferior clinical outcome in MDS, in contrast to patients with a unique hit. Strikingly, the authors did not find any difference in OS between patients who had a single hit and those with intact TP53 [15]. Moreover, Sallman et al. found that MDS patients with the TP53 somatic mutation and VAF of >40% had inferior OS (median 124 days) in comparison with those with VAF < 20% (median OS not reached) [12].
ASXL1 is mutated in 13–21% of MDS cases, and its presence was described as a predictor of inferior survival in patients with low or intermediate-1 risk according to the IPSS [10]. SF3B1 is a RNA splicing machinery member, the only gene of which the acquired lesion defines a WHO category (MDS with ring sideroblasts) [22], and its mutational status is associated with improved OS [23], independent after adjustment by IPSS-R group [11]. RUNX1 is a crucial component of normal hematopoiesis, participating in hematopoietic stem-cell genesis and differentiation. It is mutated in nearly 10% of MDS cases. Patients with a lesion in RUNX1 have significantly inferior OS compared to those without it [6][10][11][24]. Before NGS was available, it had already been proven that the presence of a RUNX1 mutation in therapy-related MDS was related to shorter time to AML, but not to impact on OS [25].
New Approaches: Machine Learning, Big Data, and “Omics” Integration
Using machine learning, Nazha and colleagues could improve the discrimination ability of current prognostic scores in the uncharted context of a post-treatment setting [26]. Whether automated algorithms, able to objectively operate with thousands of variables for a single individual, will substitute current scores mostly depends on the accuracy, size, and strict follow-up updates of training databases. International platforms were developed with the aim of gathering clinical–genetic information of patients with blood cancer into one single database from individual cases included in clinical trials and registries. The Harmony alliance, a pan-European public–private partnership, aims to reach statistical power to reveal how molecular data and treatments are intertwined.
References
- Greenberg, P.; Cox, C.; LeBeau, M.M.; Fenaux, P.; Morel, P.; Sanz, G.; Sanz, M.; Vallespi, T.; Hamblin, T.; Oscier, D.; et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood 1997, 89, 2079–2088.
- Shreve, J.; Nazha, A. Novel Prognostic Models for Myelodysplastic Syndromes. Hematol. Oncol. Clin. N. Am. 2020, 34, 369–378.
- Malcovati, L.; Germing, U.; Kuendgen, A.; Della Porta, M.G.; Pascutto, C.; Invernizzi, R.; Giagounidis, A.; Hildebrandt, B.; Bernasconi, P.; Knipp, S.; et al. Time-dependent prognostic scoring system for predicting survival and leukemic evolution in myelodysplastic syndromes. J. Clin. Oncol. 2007, 25, 3503–3510.
- Della Porta, M.G.; Tuechler, H.; Malcovati, L.; Schanz, J.; Sanz, G.; Garcia-Manero, G.; Solé, F.; Bennett, J.M.; Bowen, D.; Fenaux, P.; et al. Validation of WHO classification-based Prognostic Scoring System (WPSS) for myelodysplastic syndromes and comparison with the revised International Prognostic Scoring System (IPSS-R). A study of the International Working Group for Prognosis in Myelodyspla. Leukemia 2015, 29, 1502–1513.
- Pfeilstöcker, M.; Tuechler, H.; Sanz, G.; Schanz, J.; Garcia-Manero, G.; Solé, F.; Bennett, J.M.; Bowen, D.; Fenaux, P.; Dreyfus, F.; et al. Time-dependent changes in mortality and transformation risk in MDS. Blood 2016, 128, 902–910.
- Haferlach, T.; Nagata, Y.; Grossmann, V.; Okuno, Y.; Bacher, U.; Nagae, G.; Schnittger, S.; Sanada, M.; Kon, A.; Alpermann, T.; et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 2014, 28, 241–247.
- Makishima, H.; Yoshizato, T.; Yoshida, K.; Sekeres, M.A.; Radivoyevitch, T.; Suzuki, H.; Przychodzen, B.; Nagata, Y.; Meggendorfer, M.; Sanada, M.; et al. Dynamics of clonal evolution in myelodysplastic syndromes. Nat. Genet. 2017, 49, 204–212.
- Nazha, A.; Komrokji, R.S.; Garcia-Manero, G.; Barnard, J.; Roboz, G.J.; Steensma, D.P.; DeZern, A.E.; Zell, K.; Zimmerman, C.; Al Ali, N.; et al. The efficacy of current prognostic models in predicting outcome of patients with myelodysplastic syndromes at the time of hypomethylating agent failure. Haematologica 2016, 101, e224–e227.
- Fenaux, P.; Platzbecker, U.; Ades, L. How we manage adults with myelodysplastic syndrome. Br. J. Haematol. 2020, 189, 1016–1027.
- Bejar, R.; Stevenson, K.; Abdel-Wahab, O.; Galili, N.; Nilsson, B.; Garcia-Manero, G.; Kantarjian, H.; Raza, A.; Levine, R.L.; Neuberg, D.; et al. Clinical Effect of Point Mutations in Myelodysplastic Syndromes. N. Engl. J. Med. 2011, 364, 2496–2506.
- Papaemmanuil, E.; Gerstung, M.; Malcovati, L.; Tauro, S.; Gundem, G.; Van Loo, P.; Yoon, C.J.; Ellis, P.; Wedge, D.C.; Pellagatti, A.; et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 2013, 122, 3616–3627.
- Sallman, D.A.; Komrokji, R.; Vaupel, C.; Cluzeau, T.; Geyer, S.M.; McGraw, K.L.; Al Ali, N.H.; Lancet, J.; McGinniss, M.J.; Nahas, S.; et al. Impact of TP53 mutation variant allele frequency on phenotype and outcomes in myelodysplastic syndromes. Leukemia 2016, 30, 666–673.
- Hirsch, C.M.; Nazha, A.; Kneen, K.; Abazeed, M.E.; Meggendorfer, M.; Przychodzen, B.P.; Nadarajah, N.; Adema, V.; Nagata, Y.; Goyal, A.; et al. Consequences of mutant TET2 on clonality and subclonal hierarchy. Leukemia 2018, 32, 1751–1761.
- Pellagatti, A.; Roy, S.; Di Genua, C.; Burns, A.; McGraw, K.; Valletta, S.; Larrayoz, M.J.; Fernandez-Mercado, M.; Mason, J.; Killick, S.; et al. Targeted resequencing analysis of 31 genes commonly mutated in myeloid disorders in serial samples from myelodysplastic syndrome patients showing disease progression. Leukemia 2016, 30, 247–250.
- Bernard, E.; Nannya, Y.; Hasserjian, R.P.; Devlin, S.M.; Tuechler, H.; Medina-Martinez, J.S.; Yoshizato, T.; Shiozawa, Y.; Saiki, R.; Malcovati, L.; et al. Implications of TP53 allelic state for genome stability, clinical presentation and outcomes in myelodysplastic syndromes. Nat. Med. 2020, 26, 1549–1556.
- Malcovati, L.; Papaemmanuil, E.; Ambaglio, I.; Elena, C.; Gallì, A.; Della Porta, M.G.; Travaglino, E.; Pietra, D.; Pascutto, C.; Ubezio, M.; et al. Driver somatic mutations identify distinct disease entities within myeloid neoplasms with myelodysplasia. Blood 2014, 124, 1513–1521.
- Yoshida, K.; Sanada, M.; Shiraishi, Y.; Nowak, D.; Nagata, Y.; Yamamoto, R.; Sato, Y.; Sato-Otsubo, A.; Kon, A.; Nagasaki, M.; et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011, 478, 64–69.
- Bersanelli, M.; Travaglino, E.; Meggendorfer, M.; Matteuzzi, T.; Sala, C.; Mosca, E.; Chiereghin, C.; Di Nanni, N.; Gnocchi, M.; Zampini, M.; et al. Classification and Personalized Prognostic Assessment on the Basis of Clinical and Genomic Features in Myelodysplastic Syndromes. J. Clin. Oncol. 2021, JCO2001659.
- Menssen, A.J.; Walter, M.J. Genetics of progression from MDS to secondary leukemia. Blood 2020, 136, 50–60.
- Duncavage, E.J.; Jacoby, M.A.; Chang, G.S.; Miller, C.A.; Edwin, N.; Shao, J.; Elliott, K.; Robinson, J.; Abel, H.; Fulton, R.S.; et al. Mutation Clearance after Transplantation for Myelodysplastic Syndrome. N. Engl. J. Med. 2018, 379, 1028–1041.
- Martín-Izquierdo, M.; Abáigar, M.; Hernández-Sánchez, J.M.; Tamborero, D.; López-Cadenas, F.; Ramos, F.; Lumbreras, E.; Madinaveitia-Ochoa, A.; Megido, M.; Labrador, J.; et al. Co-occurrence of cohesin complex and Ras signaling mutations during progression from myelodysplastic syndromes to secondary acute myeloid leukemia. Haematologica 2020.
- Vardiman, J.W.; Arber, D.A.; Orazi, A.; Hasserjian, R.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M. The 2016 revision to the World Health Organization classifi cation of myeloid neoplasms and acute leukemia. Blood 2016.
- Malcovati, L.; Karimi, M.; Papaemmanuil, E.; Ambaglio, I.; Jädersten, M.; Jansson, M.; Elena, C.; Gallì, A.; Walldin, G.; Porta, M.G.D.; et al. SF3B1 mutation identifies a distinct subset of myelodysplastic syndrome with ring sideroblasts. Blood 2015, 126, 233–241.
- Harada, H.; Harada, Y.; Niimi, H.; Kyo, T.; Kimura, A.; Inaba, T. High incidence of somatic mutations in the AML1/RUNX1 gene in myelodysplastic syndrome and low blast percentage myeloid leukemia with myelodysplasia. Blood 2004, 103, 2316–2324.
- Christiansen, D.H.; Andersen, M.K.; Pedersen-Bjergaard, J. Mutations of AML1 are common in therapy-related myelodysplasia following therapy with alkylating agents and are significantly associated with deletion or loss of chromosome arm 7q and with subsequent leukemic transformation. Blood 2004, 104, 1474–1481.
- Nazha, A.; Komrokji, R.S.; Meggendorfer, M.; Mukherjee, S.; Al Ali, N.; Walter, W.; Hutter, S.; Padron, E.; Madanat, Y.F.; Sallman, D.A.; et al. A Personalized Prediction Model to Risk Stratify Patients with Myelodysplastic Syndromes. Blood 2018, 132, 793.




