1000/1000
Hot
Most Recent

Antimicrobial peptides (AMPs), also known as host defense peptides (HDPs), are produced by all living matter as a critical part of the innate immune system [1,2,3]. Their existence was discovered in 1939, the year gramicidin was isolated from the bacteria, Bacillus brevis; some resources, however, claim that the discovery of lysozyme in the 1920s should be treated as the first AMP instance, due to lysozyme’s non enzymatic, bactericidal second mode of action [2].
As of 2019, over 3000 AMPs have been isolated across all kingdoms [1]. Most of them have variable sequence length—from 10 up to 60 amino acid residues, mainly in L-configuration [1]. AMPs are expressed in many cell types in response to the activation of the Toll-like receptor (TLR) signaling pathway [2]. They are predominantly comprised of two types of amino acids residues—cationic residues such as Arg, Lys, and His, and hydrophobic residues (mainly aliphatic and aromatic) [2]. Both cationic and hydrophobic residues engage in the antimicrobial mechanism of action. Furthermore, AMPs can be classified into three groups, based on their structures (Scheme 1): α-helical, β-sheets, and extended peptides. α-helical peptides are the largest group of AMPs with characteristic qualities such as amphipathic properties, and the ability to possess a tertiary structure with a hinge in the middle of a chain [2][1]. β-sheets peptides feature one to five disulfide bridges that help to stabilize their bioactive conformation. Peptides rich in amino acids such as proline, arginine, tryptophan or glycine usually have a linear structure [2][1].
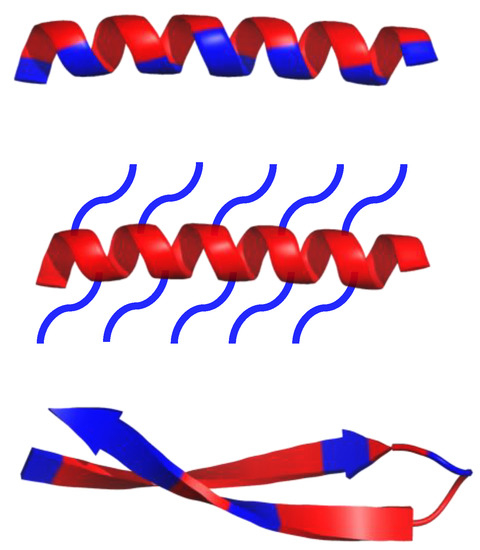
Scheme 1. Schematic representation of α-helical and β-sheets, common secondary structure of amphipathic peptides. Hydrophobic amino acid residues are colored red, while hydrophilic residues are colored blue.
HDPs have a wide range of antibacterial activities, and low susceptibility to antimicrobial resistance [5][6][7][8][9][10][11][12]. Nevertheless, the use of HDPs is limited due to their low resistance to proteases, and their increased cost of preparation [8][13][14]. Different peptides [15][16][17][18], peptoids [19][20][21][22][23], polymethacrylate derivatives [24], polynorbornene derivatives [25], polycarbonate derivatives [26], peptide polymers [27][28][29][30][31][32], and polymers made by controlled living radical polymerization (CLRP) [33] were developed to mimic the antibacterial properties of HDPs, and to ameliorate their shortcomings [34].
The widely developed peptide-drug research has led to the introduction of around 60 FDA approved peptide therapeutics in the market [35]. Over 140 are in clinical trials, and over 500 are in the preclinical stages. Although numerous biomimetic and de novo designed AMPs are at different stages of clinical trials [36][37], only eight AMPs received FDA approval, i.e., daptomycin (approved in 2003) and oritavancin (approved in 2014) [37].
There are three main factors—charge, hydrophobicity and amphipathicity, which determine the activity of AMPs, and they are all related to their intrinsic properties.
The positive charge of AMPs (between 2+ and 9+) is one of the most essential factors responsible for their antibacterial activity [38]. Positively charged AMPs outcompete the binding of native Mg(II) and Ca(II) ions to lipopolysaccharides (LPS), and destabilize the outer membrane of Gram-negative bacteria [39]. Consequently, the destabilized regions enable the peptide to pass into the cell. Once through, AMPs bind to the cytoplasmic membrane and cause depolarization and pore formation, resulting in cell death [40]. In the case of Gram-positive bacteria, AMPs must overcome the outer wall with two main obstacles, peptidoglycan and teichoic acids, before an AMP interacts with the cytoplasmic membrane. AMPs overcome peptidoglycan because it is able to penetrate through the relatively porous peptidoglycan. The porous nature of peptidoglycan makes it easy for small molecules (< 50 kDa) to penetrate through it, and furthermore, it does not possess a negative charge, which could prevent the penetration of positively charged AMPs through it [40][41][42]. To overcome the anionic teichoic acids, they can either act as a ladder that will help positively charged AMPs to travel to the cytoplasmic membrane, or the can act as “cages” that entrap AMPs inside the bacteria, thus reducing their local concentration on the membrane The behaviors of teichoic acids vary based on the type of AMP and the type of bacteria [42]. After crossing the outer wall, AMPs can disrupt the integrity of the cytoplasmic membrane, leading to membrane disruption and dislocation of peripheral membrane proteins [42].
The hydrophobicity of AMPs is another important factor. It is assumed that hydrophobic residues help to insert peptides into the bacterial membrane and further impair the membrane [4][42]. It was also revealed that hydrophobic residues account for 40–60% of all amino acids in AMPs [38]. Moreover, the hydrophobicity is strongly correlated with antimicrobial activity, where increasing the hydrophobicity to a certain level improves antimicrobial activity [1][38]. The relationship between hydrophobicity and antimicrobial activity is also proven to be the case for peptidomimetics such as peptoids, where amino acid side chains are linked to the peptide backbone through the amide nitrogen rather than to the α-carbons [43]. However, a higher hydrophobicity also increases hemolytic activities, resulting in unwanted toxicity to eukaryotic cells [1][38]. Studies with model peptides have shown that a continuous region of 4-6 hydrophobic residues is sufficient to sustain the antimicrobial activity of peptides, while simultaneously reducing the hemolytic activity [44].
The amphipathicity of AMPs, created by the segregation of hydrophobic and polar residues on the opposite sites of the backbone, is another factor related to their activity [1]. Amphipathicity is considered the strongest indicator of AMP activity, and is described by the hydrophobic moment, defined as the vector sum of the hydrophobicity of each amino acid located in the helix [1][45]. The mechanism by, which amphipathicity works is the result of previous factors. The positively charged regions of AMPs enable binding to the anionic phospholipid head groups of the bacterial membrane. Meanwhile, the hydrophobic regions of peptides invade the lipid bilayer and interact with the hydrophobic acyl chains of phospholipids, which results in membrane penetration [45]. Several studies have shown the importance of amphipathicity in the process of AMPs binding to membranes, and the negative impact on activity against Gram-positive bacteria and fungi, when amphipathic character of the peptide was eliminated [13][46][47][48].
Despite the recent advancements in AMP research, there are still major challenges to overcome. One of the main obstacles in AMP applications is the proteolytic instability of peptide drugs [49][50]. Peptides are targeted by numerous proteolytic enzymes located in bodily fluids and tissues of the host. Moreover, AMPs can be recognized as an antigen and targeted by host immune system [51]51[52]. One of the countermeasures for their potential immunogenicity could be glycosylating AMPs with a glycosyl profile similar to that of the host. For instance, covering AMPs with polysialic acid (derived from Neisseria meningitidis or E. coli that shares structural mimicry with host cell lectins), or its analogues, can lessen the immunological response [52]. Nevertheless, some bacteria have already developed resistance to AMPs. For example, LL-37, a human antimicrobial peptide, can be digested by proteases synthesized by P. aeruginosa, Enterococcus faecalis, Proteus mirabilis, S. aureus, and S. Pyogenes [50][53]. Synthesizing proteases to target specific antimicrobial peptides is one of the many ways bacteria participate in this evolutionary “arms race”. Other methods used by bacteria to cope with AMPs, including the use of efflux pumps, lowering the binding affinity, or reducing the anionic charge of lipopolysaccharides on their surface, are exhaustively discussed in [50][53].
The high toxicity of AMPs in eukaryotic cells is another one of their shortcomings. This high toxicity can lead to hemolysis, nephrotoxicity, and neurotoxicity [1][55][56][57]. More studies need to focus on the pharmacokinetics and pharmacodynamics of AMPs in order to determine a proper drug dosage that will maintain a balance between positive and negative outcomes [2].
The bioavailability of AMPs is rather low. Peptides are hardly absorbed by the intestinal mucosa, and their pharmacological distribution requires additional financial input. In recent years, much progress has been made in this field—e.g., nano-carriers not only increase the bioavailability, but also lower cytotoxicity and reduce degradation of a compound, leading to increased efficiency [37]. Despite different efforts, the entire process of AMPs design and discovery (costs of synthesis and screening) is relatively high and does not guarantee the expected outcome [58]. The high production cost of AMPs (as for 2006, producing 1 g of peptide using solid-phase peptide synthesis (SPPS) costs USD 100–600) limits the development and testing of new AMPs, even if recent improvements in SPPS lowered the costs of synthesis [8].
AMPs, as other peptide drugs, can induce immune responses and cause allergies [59]. Immunogenicity depends on the host immune system, but is also correlated with the peptide’s dose, duration and frequency of treatment, route of administration, and the patient’s pathologies [60][61][62][63]. Immunogenic response can affect the peptide-drug efficiency and even lead to allergy and/or hypersensitivity [63]. Bering that in mind, it is important to study the immunogenic and allergic activity of AMPs. In vitro tests together with mathematical calculations of allergens similarities, with the use of specific databases of allergenic compounds [64], i.e., http://www.allergome.org, can give nowadays reliable results [65].