1000/1000
Hot
Most Recent

Pectobacterium brasiliense (Pbr) is a worldwide-distributed bacterial plant pathogen causing soft rot of a wide range of economically important crops.
The group of bacterial plant pathogens known as pectinolytic, soft rot Pectobacteriaceae (SRP) consists of two genera: Pectobacterium and Dickeya [1]. These pathogens are responsible for plant tissue maceration resulting in water-soaked lesions which lead to collapse of the infected tissue, wilting and death of the plants [2][3]. Pectobacterium brasiliense (Pbr) is a worldwide-distributed bacterial plant pathogen causing soft rot of a wide range of economically important crops. Pbr has been reported in Eurasia: from Western European countries to Russia and China [4][5][6], in Africa: South Africa, Morocco, Algeria and Kenya [7][8][9][10]. Pbr has been constantly reported since 2004 causing significant losses particularly of potato (Solanum tuberosum L.) [11][12][13][14] and posing a threat to worldwide potato gross production value which is estimated to be $63914 million in 2018 [15]. Significant disease caused by Pbr has been pointed out in Belgium, the Netherlands and Switzerland: for instance, in 2015 in Switzerland prevalence of Pbr reached 70% of the samples collected from potato field outbreaks [13]. In Russia, in 2017 more than 20% soft rot disease was caused by Pbr and this species also has been reported as a predominant cause of blackleg of potato in the Moscow region in 2018 [14]. However, in some other countries such as the United Kingdom and Norway, so far Pbr is not considered as a problematic species in potato fields [16][17][18]. In addition to losses in potato production, Pbr causing loses of other crops: for instance, in the years 2014–2015, in five Chinese provinces (Shandong, Shanxi, Hebei, Henan, and Liaoning), Pbr caused soft rot of cucumber whereby the disease incidence was vary from 15 to 50% in different fields, causing 20 to 30% yield losses [6]. Hence, Pbr is considered to be a highly aggressive pathogen and is causing more soft rot/blackleg disease outbreaks elsewhere than other Pectobacterium species [4][19].
During the last two decades, high-throughput sequencing (HTS) technologies have been an important driving force in the progress of life sciences [20]. Genomic information has also been one of the cores of molecular biology in studying evolutionary biology, taxonomy, phylogeny, and it can also provide insights to distinct niches adaptation [21][22]. The availability of several published genomes illuminates new strategies for exploring the genomic datasets, through comparative genomics approaches. Therefore, some genomic comparisons have also allowed for delineating taxa, for identifying the core repertoire of virulence genes shared by related organisms and, for locating gene clusters or genome islands exclusive of species or even unique of a strain for developing molecular tools for identification and detection of pathogens [23]. To date, few comparative studies have also been conducted regarding Pectobacterium species [24][25][26][27][28]. Until now, six Pbr complete genome sequences are publicly available: Pbr PCC21 (misnamed as P. carotovorum PCC21 in NCBI) isolated from B. rapa ssp. pekinensis [29], Pbr BZA12 from Cucumis sativus [25], Pbr BC1 from Brassica rapa ssp. pekinensis [30], Pbr SX309 from C. sativus fruit [31], Pbr HNP201719 and Pbr 1692 from S. tuberosum [32]. In addition, more than thirty draft genomes of Pbr species have also been deposited in the GenBank genome database.
Over the past decades, the taxonomy of the genus Pectobacterium has undergone major modifications (Figure 1) [33][34][35][36][37][38][39][40][41][42][43][44][45][46][47][48]. Establishment of Pbr as a separate taxon was initially based on amplification of the intergenic spacer region in PCR, differences in 16S rRNA gene sequence, and analysis of biochemical traits [11]. Later, it was proposed as a subspecies of P. carotovorum through multi-locus sequence analysis (MLSA) in several studies [49][50]. This taxon has been recently elevated at a species level Pbr using a combination of phylogenomics, in-silico DNA-DNA hybridization (isDDH) (70% threshold), average nucleotide identity (ANI) (95–96% threshold) and biochemical traits [34]. In the same paper, the authors also described P. odoriferum, P. actinidiae and P. versatile as new species, and emended description of P. carotovorum. Because of this recent change, we have used the updated species description P. carotovorum, but we alert the readers that some of these P. carotovorum strains may potentially belong to P. versatile.
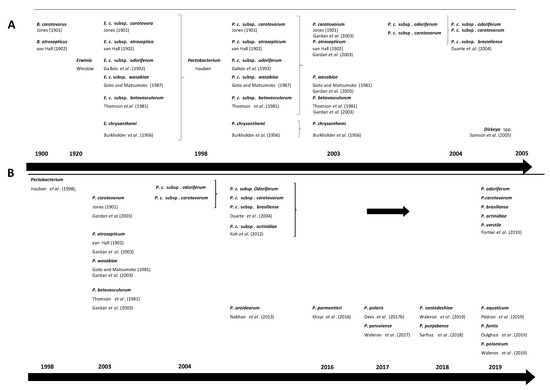
Figure 1. Timeline changes in taxonomy of Pectobacterium spp. (A) changes from early described species causing soft rot symptoms Bacillus carotovorus and Bacillus atrosepticus, through establishment of Erwinia species by Winslow et al. up to re-classification of Pectobacterium and Dickeya spp. by Samson et al. [11][33][40][46][47][51][52][53][54][55] (B) changes from re-classification of Pectobacterium spp. up to most recent proposed species [11][33][34][38][46][47][51][52][55].
Pbr species are diverse and less homogeneous than other Pectobacterium spp. Indeed, the ANI Pbr species threshold is 95% while the ANI species threshold is often 96% for other Pectobacterium spp. Aside from improvement in Pbr species delineation, comparative genomics gives an insight into gene richness in the Pbr genomes. The analysis of the Pan-genome and Core-genome of 30 Pbr strains was conducted using the software Proteinortho V6 [56]. The genes were clustered based on the criteria of 50% identity on at least 50% of the length of the alignment. The analysis highlighted an accumulative number of 8210 gene families within the 30 strains analyzed (Figure 2A).
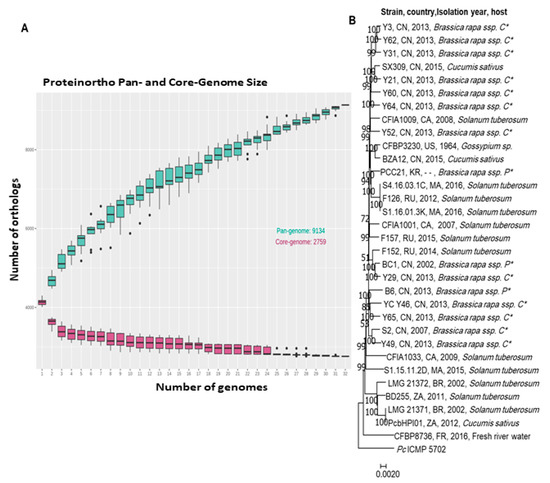
Figure 2. The genomic comparison of the available Pbr genomes in NCBI: (A) core and pan genome analysis, (B) phylogenomic analysis of the concatenated core genes of 30 Pbr strains was conducted using Proteinortho V.6 [56]. The genes were clustered based on the criteria of 50% identity on at least 50% of the length of the alignment, the P. carotovorum ICMP5702 was used as out of the group. (P *: Pekinensis; C *: Chinensis).
The pan-genome curve indicates an exponential increase in size over adding new strains stating an open pan-genome. Based on this result, we could predict an expansion of the unique gene pool by the addition of new Pbr genomes. In contrast, the pan-genome, gene number in the core-genome decreased upon addition of new strains to reach a total of 2968 core genes. Remarkably, the phylogenetic tree generated from concatenated core-genome genes showed no clear congruence between the evolutionary relationships of the Pbr isolates and the geographical origin or plant host of their isolation (Figure 2B). The relationships between the genome diversity of Pbr isolates and aggressiveness on plant hosts should be experimentally investigated using plant assays.
Comparative genomics revealed highly conserved virulence genes in the Pectobacterium [28]. Some of these virulence factors include, amongst other quorum-sensing (QS), secretion systems, adhesion, plant cell wall-degrading enzymes (PCWDEs), motility, chemotaxis, siderophores and biofilm formation [30][31][57][58][59]. Similarly, knowledge of the phylogeny and the genetic basis for the pathogenicity of Pectobacterium was expanded recently, as 84 Pectobacterium genomes were screened for the presence of 159 genes that are known as virulence factors [30]. Only a few studies have focused on studying the complete genome of Pbr, and consequently, the pathogenicity and the mechanisms for genetic adaptation to the host remains largely unknown [31]. The genomic analysis of Pbr (reported as P. carotovorum subsp. brasiliense SX309) showed the presence of many similar virulence factors already described in Pectobacterium spp. including the PCWDE biosynthetic genes, bacterial QS genes, secretion system genes, chemotactic genes and flagella [28][31]. In Pbr SX309 several variable regions of two subtype CRISPR-Cas immune systems and type VI secretion system may also contribute in the infection process [31]. A study conducted by Moleleki et al. [58] showed that Pbr colonization, swimming motility, and flagella biosynthesis are also regulated by the QS system. In addition, Pbr 1692 is known to be more aggressive and typically outcompetes other members of SRP [60]. The production of several antimicrobial compounds by Pbr 1692 could contribute to its capacity to effectively colonize different types of ecological niches [61].
Bacteria belonging to the genus Pectobacterium were considered among the most threatening of phytopathogens to the health of vegetable, ornamental and fruit crops, including Pbr [11][62]. Pbr is responsible for the degradation of the cell wall of several plant hosts and causing blackleg and soft rot diseases (Figure 3).
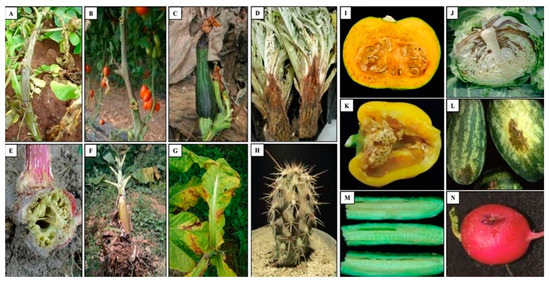
Figure 3. Symptoms caused by Pbr in several plant species. (A) Potato (S. tuberosum), (B) Tomato (Solanum lycopersicum), (C) Zucchini (Cucurbita pepo), (D) Artichoke (Cynara cardunculus var. scolymus), (E) Amaranth (Amaranthus), (F) Banana (Musa sp.), (G) Tobacco (Nicotiana tabacum), (H) Tetecho (Neobuxbaumia tetetzo), (I) Squash (Cucurbita pepo), (J) Cabbage (Brassica oleracea), (K) Pepper (Capsicum annuum), (L) Watermelon (Citrullus lanatus), (M) Cucumber (Cucumis sativus), (N) Raphanus (Raphanus sativus) [6][7][63][64][82][85][86][87].
Indeed, Pbr infects a wide range of plant species, including both monocotyledon and dicotyledon clades [7][11][62][63][64][65]. Nowadays, 19 different plant species belonging to 10 families are reported as hosts of Pbr (Table 1). As Pbr was isolated in different continents, it has apparently adapted to many environments and climates, including tropical and temperate regions. Therefore, this pathogen could be more widespread than currently known.
Table 1. Hosts range, disease symptoms and geographical distribution of Pbr.
| Clade | Family | Host (Plant Species) | Region/Country | General Disease | Symptoms |
|---|---|---|---|---|---|
| Dicotyledons | Solanaceae | Potato (Solanum tuberosum) | Brazil, Kenya, Japon, Canada, South Africa, Switzerland, Poland, New Zealand, South Korea, Netherlands, Algeria, Turkey, Russia, China, Egypt, USA, Hawaii, Thailand, Morocco, Zimbabwe, Syria, France | Soft rot and blackleg | The infected plants were stunted with yellowish foliage, water-soaked regions with watery ooze, darkened and necrotic basal stem symptoms extending upward. |
| Tomato (Solanum lycopersicum ) | South Korea, Colombia, USA, Italy | Stem rot | Soft and aqueous lesions, dark brown discoloration of the basal part of stem petioles, water-soaked pith tissue and internal necrotic. | ||
| Pepper (Capsicum annuum) | Venezuela, South Korea, China | Soft rot and black spot | Watery lesions at the basal part of the stem, water-soaked and necrotic tissue, defoliation and fruit decay. | ||
| Eggplant (Solanum melongena) | South Korea | Soft rot | Water-soaked lesions, soft rot symptoms on fruits, discoloration of vascular tissues, stem hollowness, and dark green lesions that turned brown with age. | ||
| Tobacco (Nicotiana tabacum) | China | Bacterial leaf blight | Necrosis along the main or lateral veins, drying and rotting of the leaves. | ||
| Cucurbitaceae | Cucumber (Cucumis sativus) | China, South Africa | Soft rot | The gummosis emerged on the surface of leaves, stems, petioles, and fruit. The basal stem color was dark brown and the stem base turned to wet rot. Yellow spots and wet rot emerged at the edge of the infected leaves and gradually infected the leaf centers. | |
| Zucchini (Cucurbita pepo) | Poland, Brazil, Serbia, Austria, Italy | Soft rot | Round water-soaked lesions. The affected tissues were light brown, slightly sunken, soft, and macerated. | ||
| Watermelon (Citrullus lanatus) | Serbia | Soft rot | Soft rot brownish lesions of stems, collapse and wilting of entire vines. | ||
| Brassicaceae | Cabbage (Brassica oleracea var. capitata) | Poland | Soft rot | Symptoms were characterized by gray to pale brown discoloration and expanding water-soaked lesions on leaves. | |
| Chinese cabbage (Brassica rapa ssp. pekinensis and chinensis) | South Korea | Soft rot | Water-soaked lesions, affected tissue turns brown and becomes soft and mushy. Leaves, stems, and roots may decay entirely. | ||
| Raphanus (Raphanus sativus) | China | Root rot | The infected plants were stunted with yellowish foliage and blackened center leaves and the infected roots exhibited a completely decayed pith region. | ||
| Asteraceae | Chrysanths (Chrysanthemum) | South Korea, France, USA | Soft rot | * | |
| Artichoke (Cynara cardunculus var. scolymus) | Italy | Soft rot | Chlorosis and wilting of the older leaves accompanied by dark-green to dark-brown soft rotting of the pith. | ||
| Amaranthaceae | Sugar beet (Beta vulgaris) | Poland, USA | Soft rot | Soft decay of internal root tissues, reddening of affected tissue after cutting, blackening of petiole vascular bundles, half-leaf yellowing, and frothing. | |
| Chenopodiaceae | Amaranth (Amaranthus) | South Korea | Soft rot | Wilting, defoliation and odd smell. | |
| Cactaceae | Tetecho (Neobuxbaumia tetetzo) | Mexico | Soft rot | Soft rot that damages the whole plant and causes its fall and disintegration. | |
| Nepenthaceae | Nepenthes (Nepenthes) | South Korea | Soft rot | * | |
| Malvaceae | Bull Mallow (Malva nicaeensis) | Israel | ** | ** | |
| Gossypium sp. | USA | * | * | ||
| Primulaceae | Cyclamen sp. | France | * | * | |
| Caricaceae | Carica papaya | France (Overseas territory, La Réunion) | * | * | |
| Monocotyledon | Musaceae | Banana (Musa sp.) | India, France (Overseas territory, Martinique) | Rhizome rot | Massive soft rot accompanied by disagreeable foul-smelling rot of the rhizome and internal decay of the pseudostem as the infection spread upward. |
| Non-Host Environment | Water | Spain | * | * | |
| Rhizosphere of Solanum dulcamara | France | * | * | ||
* Symptoms were not described, ** Symptomless.
The family Solanaceae is considered as the major host of Pbr. Five host plant species belonging to this family have been reported so far, including potato (S. tuberosum L.), tomato (S. lycopersicum), pepper (C. annuum), eggplant (S. melongena), and tobacco (N. tabacum) (Table 1). On the basis of available studies reporting the presence of Pbr in the host plants, more than 50% of them have been associated with potato (this study, Table 1). In fact, Pbr has been reported in potato in Brazil since 2004 [11], followed by its detection in North America [66,67,68], Europe [12][13][14][69][70], Africa [7][9][71][72], Asia [62][73][74][75][76], and New Zealand [77]. Generally, Pbr causes blackleg and soft rot in potato leaves, stems, and tubers [8][10][66][68][74]. Tomatoes have been reported to be also infected by Pbr, resulting in a large loss for producers. In Colombia, Jaramillo et al. [78] reported the presence of Pbr in ‘Calima’ hybrid tomato, causing aqueous and brown lesions on the lower stem, necrosis of the vascular bundles, and some plants presented cracking symptoms along the stem. In Florida, Pbr infected “heirloom” tomatoes, causing wilting, necrosis of leaves and stems, and water-soaked pith tissue [79]. Pbr caused brown water-soaked, soft-rotted pith tissue, and internal vascular discoloration in Grafted tomato plants from Sicily (Italy) [80]. In the pepper, Pbr caused water-soaked, necrotic tissues, and wilt with defoliation in Venezuela [81], fruit decay and pedicel decay in Korea [82], and black spot disease in China [83]. Pbr causes soft and aqueous legions in eggplant from South Korea [62], as well as necrosis, drying and rotting in the leaves of Tobacco from China [84].
The plant families Cucurbitaceae and Brassicaceae are also affected by Pbr. Infection of cucumber was reported in China [6] and South Africa [90], and was characterized by the appearance of gummosis on the surface of leaves, stems, petioles, and fruits, dark brown coloration of the basal stems, as well as yellow spots could sometimes emerge at the edge of infected leaves [6]. The infection of zucchini were reported in Poland [69], Serbia [65], Austria [91], Brazil [92], and Italy [88], and was characterized by water-soaked lesions and fruits macerating. Infection of squash and watermelon were reported in Northern Serbia [65] with disease appearing in squash as light brown, slightly sunken, soft, and macerated tissue with the presence of a water-soaked lesions. Infection caused by Pbr in watermelon developed as a soft rot brownish lesions on the infected stems. Furthermore, soft rot in cabbage and Chinese cabbage were reported in Poland [69], and South Korea [62], characterized by water-soaked lesions on leaves and gray to pale brown discoloration of tissues. However, root rot disease of Raphanus was reported in China [93], in which the infected plants were characterized by yellowish foliage, blackened center leaves, and decayed roots.
Asteraceae and other host families like Amaranthaceae, Chenopodiaceae, Cactaceae, Nepenthaceae, Malvaceae, Primulaceae, and Caricaceae were rarely infected by Pbr. In Sicily (Italy), the infected Artichoke (Cynara cardunculus var. scolymus) develops wilting of the older leaves accompanied by dark-green to dark-brown soft rotting of the pith [63]. In a study carried out in the USA, infected sugar beet (B. vulgaris) was characterized by soft decay of internal root tissues, blackening of petiole vascular bundles, half-leaf yellowing, and frothing [94]. Moreover, Amaranth (Amaranthus L.) grown in South Korea has been infected by Pbr presenting typical soft rot symptoms like wilting, defoliation and odd smell [86]. In Mexico, the infection of Tetecho (N. tetetzo) by Pbr causes damages of the whole plant, as well as collapse and disintegration [85]. Pbr is also known to cause asymptomatic infections in some plant species.
Recently, a survey was conducted in potato fields where the infected plants were detected, showing the isolation of Pbr from asymptomatically infected Malva nicaeensis [95]. However, the mechanism of infection in asymptomatic hosts is not well understood.
Soft rot diseases caused by Pbr are rare in monocotyledon hosts. Only one study has reported the presence of Pbr in hosts belonging to monocotyledon clade. This study isolated the pathogens from different cultivars of banana (Musa sp.) in India and the French overseas territory Martinique [64][88], in which the pathogen causing rhizome rot was characterized by disagreeable foul-smelling and internal decay of the pseudostem [64].
Although Pbr can infect plants from both monocotyledons and dicotyledons, it can present some specialization toward the host at the strain level. In fact, nine strains of Pbr isolated from cucumber were in vitro tested against different plants species. These strains caused soft rot in potato, tomato, green pepper, broccoli, radish, mustard, zucchini, cucumber and others, but did not cause disease in balsam pear and loofah [6]. In another study, strains of Pbr isolated from N. tetetzo caused soft rot on many plant species, including Myrtillocactus geometrizans, Opuntia ficus-indica, S. lycopersicum, Cucumis sativus, and Daucus carota subsp. sativus, but they did cause symptoms on C. pepo, Physalis ixocarpa, and Brassica oleracea var. capitata [85].
Moreover, Pbr has been isolated from non-host environment, including water in Spain and the rhizosphere of S. dulcamara in France [34][88]. Pbr is considered a major threat for horticulture crops in these regions, as can be easily transmitted to other fields through irrigation water systems or by its persistence in the soil.
Visual assessment of blackleg and soft rot disease symptoms of infected plants is not enough to confirm the presence of Pbr, as the symptoms are indistinguishable from infection caused by other SRP. Hence, the use of accurate detection and identification tools is required to study the biodiversity and pathogenicity of Pbr.
Symptomatic plants should be collected and stored in a cool container until they are received in the lab, and their diseased parts processed to isolate SRP. Naas et al. [9] collected tuber samples from the margins of diseased tissue of potato, and after sample maceration in sterile water, streaked material on nutrient agar plates, incubated at 27 ± 1 °C. Gillis et al. [81] used fruits, stems and leaves from infected pepper (C. annuum L.) to isolate pathogens while other microbes were eliminated by surface sterilizing tissues for 30 s in 75% ethanol. Other researchers have used 0.6% sodium hypochlorite (NaOCl) for 30 s [96], 0.6% NaOCl for 1 min [97], and 70% ethanol for 30 s followed by 0.5% NaOCl for 30 s [83]. The streaked agar plates are incubated at optimal temperatures (26–28 °C) for Pbr growth for different incubation times (48 to 78 h) depending on the medium.
Several non-selective media are used for the culture of Pbr, such as nutrient agar, tryptic soy agar and Luria-Bertani agar [62]. Besides, other semi-selective-diagnostic media can be used in order to detect, isolate and enumerate SRP. One of the most used media for this purpose is crystal violet pectate (CVP) medium and its modifications [98][99]. This is a semi-selective medium that contains pectin as a carbon source and crystal violet as an inhibitor for the growth of Gram-positive bacteria. SRP are secreting PCWDEs, which metabolize pectin, resulting in formation of characteristic cavities in this solid medium [100]. From its first use by Cuppels and Kelman [101], this medium was modified and improved in order to provide a better pectin source that can be easily degraded by bacteria. For example, Hélias et al. [100] tried six different pectin sources. Among them, they found that AG366 pectin was highly effective in CVP medium compared to commercial ones. The characteristic of this medium makes it suitable for identification of SRP. It provides better results as it allows formation of deep cavities due to degradation of pectin by microbes [100].
The most of biochemical tests are used as additional methods for confirmation of identity and genetic grouping of Pectobacterium including Pbr [9]. Biochemical assays used for identification and differentiation of Pbr including Gram staining, the ability of isolates to produce oxidase and catalase [85], carbon source utilization [34][102], acid production from maltose and α-methyl-D-glucoside, growth at 29 °C and 37 °C, tolerance to 5% NaCl, erythromycin sensitivity, indole production, lactose fermentation, erythromycin sensitivity and gas production from D-glucose [6]. The Hugh and Leifson medium was employed to study the oxidative or fermentative metabolism of glucose [68]. Furthermore, King’s B medium was used to evaluate the ability of Pbr isolates to produce fluorescent pigments [81]. One of the commercial tests available for this purpose is GN2 microplate which is designed for the characterization and identification of different aerobic Gram-negative bacteria including Pbr [6][62].
Several molecular method have been employed for the characterization of Pbr, including conventional polymerase chain reaction (PCR) [8][9], real-time qPCR [103], PCR-restriction fragment length polymorphism (PCR-RFLP) [10], pulsed-field gel electrophoresis (PFGE), MLSA [62] and HTS [8], with conventional PCR and real-time PCR commonly used in laboratories for robust confirmation of Pbr in the samples [9][103] (Table 2).
Table 2. Isolation and detection methods of Pbr.
| Classification | Method | Applications | Primers/Probes | Features | Target Species |
|---|---|---|---|---|---|
| Artificial media | CVP, modified CVP (single or double layer), enrichment using PEB, other formulations | Isolation pure cultures of bacteria | Pectinase | Degradation of polypectate or other reaction | Pectobacterium and Dickeya spp. |
| PCR methods | Conventional | Identification to genus level | Y1 & Y2 | pel gene | Pectobacterium spp. including Pbr |
| Identification to species level | BR1f/L1r | 16S–23S rRNA | Pbr | ||
| Real-time (qPCR) | Identification and quantification | Pb1F/Pb2R; Probe name PbPr | (16S-23S ITS) and tRNA-Glu gene AraC sequences |
Pbr | |
| PbrFW/PbrRv; Probe name Pbrb | |||||
| DNA Sequencing methods | Single gene sequencing | Identification of species | gapA326F/gapA845R | gapA gene | Pectobacterium and Dickeya spp. |
| mdh86F/mdh628R | mdh gene | Pectobacterium spp. | |||
| Pec.dnaA-F1/Pec.dnaA-R1 | dnaA gene | Pectobacterium spp. | |||
| Whole genome sequencing (MLSA, ANI, isDDH, phylogenomics) | Identification of species | Not applicable | Not applicable | The study of phylogenetic relationships of species within a genus |
The most recommended method for differentiation of Pbr from other SRP are MLSA and HTS enable to confirm phylogenetic position within Pectobacterium genus [8][34]. Additionally, among other reported detection methods (Table 2), MLSA is widely used due the availability of the data in the public data base (NCBI). Also, it was used for taxonomic information [34][111] and the study of the diversity of Pbr with the related species [8][9]. Most recently, several housekeeping genes were reported to differentiate Pbr in MLSA schemes [8][9][34][50]. For example, Nabhan et al. [50] have used eight housekeeping genes in MSLA to delineate species of Pectobacterium, including glutamylphosphate reductase (proA), aconiate hydrase 1 (acnA), mannitol-1-phosphate 5-dehydrogenase (mtlD), isocitrate dehydrogenase (icdA), malate dehydrogenase (mdh), glucose-6-phosphate isomerase (pgi), glyceraldehyde-3-phosphate dehydrogenase A (gapA), and the RNA polymerase subunit sigma factor 38 (rpoS). However, as these methods highly efficient for differentiation of Pbr are time consuming and need qualified personnel and specialized equipment to perform analysis.
In contrast to other widely distributed SRP (i.e., P. atrosepticum, P. carotovorum and Dickeya) any of sensitive and robust POCT detection methods such as isothermal amplification or handled biosensor device have not been developed thus far as specific for detection of Pbr. These reported POCT methods are specific for P. atrosepticum [112], P. carotovorum [113], P. aroidearum [114] or Dickeya spp. [115][116][117]. However, fairly recently interesting approach has been reported by Ahmed et al. [118] for detection of Pectobacterium spp. including Pbr. Developed assay based on recombinase polymerase amplification adopted on lateral-flow device (RPA-LFD) was reported as sensitive (10 fg/μL) and specific handled device detecting Pectobacterium spp. including Pbr. Additionally, the benefits coming from the use of this POCT method is performing a test directly from infected plant material with no need for DNA extraction [118]. Recently, the use of infrared spectroscopy and machine learning has been proposed as a cheaper (relative to molecular techniques) method of identifying and differentiating between different genera, species and strains of SRP [119].