1000/1000
Hot
Most Recent

Selective autophagy is a highly regulated degradation pathway for the removal of specific damaged or unwanted cellular components and organelles such as protein aggregates. Cargo selectivity in selective autophagy relies on the action of cargo receptors and adaptors. In mammalian cells, two structurally related proteins p62 and NBR1 act as cargo receptors for selective autophagy of ubiquitinated proteins including aggregation-prone proteins in aggrephagy. Plant NBR1 is the structural and functional homolog of mammalian p62 and NBR1.
Autophagy is an evolutionarily conserved process in eukaryotic organisms for degradation of cytoplasmic constituents including proteins and organelles in the lysosomes or vacuoles [1]. There are at least three types of autophagy, known as macroautophagy, chaperone-mediated autophagy and microautophagy. Macroautophagy is the pathway that has been most extensively characterized and is often referred to simply as autophagy [1]. Autophagy is usually induced in response to a variety of physiological and environmental stimuli and plays an important role in cell homeostasis under unfavorable and pathological conditions including starvation, extreme temperature, aging and pathogen infection [2]. Induction of autophagy is initiated by the formation of an isolation membrane called phagophore that can expand to capture cytoplasmic components and enclose the sequestered cargos to form a separate compartment, the double-membrane autophagosome [3][4]. The autophagosome is then primed to fuse with the lysosomes or vacuoles for degradation of captured materials by lysosomal/vacuolar hydrolases. Autophagosome biogenesis is a complex membrane process that requires more than 40 autophagy-related (ATG) proteins. These ATG proteins function in several physiologically continuous, but mechanistically distinct steps and are organized as several functional groups, which include (i) the ATG1–ATG13–ATG17 scaffold, formed upon activation of the ATG1 kinase during the early stage of autophagy induction, (ii) the class III phosphatidylinositol 3-kinase (PtdIns3K)–ATG14–ATG6 (Beclin 1) complex for the nucleation and assembly of the initial phagophore membrane, and iii) two interrelated ubiquitin-like conjugation systems, ATG12–ATG5–ATG16 and ATG8–PE (phosphatidylethanolamine), which are required for the regulation of membrane elongation and expansion of the forming autophagosomes [1].
The core process of autophagy and ATG proteins are highly conserved in all eukaryotic organisms including plants [1][5][6][7]. Using genetic and molecular approaches, extensive studies over the past two decades or so have demonstrated an important role of autophagy in almost all aspects of plant life, particularly in plant stress responses [5][8][9]. Autophagosome biogenesis and ATG gene expression are both induced by diverse abiotic stress conditions including nutrient starvation, heat, salt, drought and oxidative stresses [10][11][12][13][14][15]. Autophagy mutants and transgenic silencing lines are hypersensitive to nutrient starvation and compromised in tolerance to these abiotic stresses [10][11][12][13][14][15]. In addition, autophagy plays an important role in plant innate immunity. Plant mutants or transgenic silencing lines for autophagy are altered in responses to virulent and avirulent biotrophic pathogens including pathogen-induced hypersensitive responses [16][17][18][19][20][21]. Autophagy deficient mutants are compromised in resistance to necrotrophic pathogens [17][22]. As will be discussed below, autophagy is also involved in plant interaction with viral pathogens through such mechanisms as regulation of antiviral RNA silencing and targeting degradation of viral proteins. Autophagy also plays important roles in plant growth and development including root growth, leaf senescence, pollen and endosperm development [8][20][23][24][25].
Even though autophagy was initially considered to be a nonselective process of bulk degradation of intracellular contents, it has been now well established that the broad roles of autophagy are primarily mediated by selective clearance of specific cellular structures [26][27][28][29]. Ubiquitin-like ATG8, which is required for phagophore initiation, elongation and maturation, also plays a critical role in selective autophagy [27]. Upon attachment of the lipid PE to its carboxyl terminus through a conjugation pathway, ATG8 is anchored in both the inner and outer membrane of autophagosomes and provides a docking platform for the selective recruitment of cargos through selective autophagy receptors [27]. Most selective autophagy receptors recognize specific cargos and also interact with membrane-anchored ATG8 through ATG8-interacting motifs (AIMs), which have the W/Y/F-X-X-L/I/V consensus core sequence [27]. The AIM motif binds a hydrophobic patch on ATG8 known as AIM docking site [27]. Recently, a new class of selective autophagy receptors have been identified, which interact with ATG8 through ubiquitin-interacting motif (UIM)-like sequences for high-affinity binding to an ATG8 interaction site different from the AIM docking site [30][31]. Through assays with candidate UIM-containing proteins and unbiased screens, a large number of UIM-based ATG8 interactors have been identified in plants, yeast, and humans [32]. Discovery of the new class of selective autophagy receptors greatly expands the scope of selective autophagy [32].
A large number of autophagy receptors from nonplant organisms have been identified that mediate the selective autophagic degradation of a wide range of cargoes including protein aggregates, signaling complexes, mitochondria, peroxisomes, endoplasmic reticulum (ER), ribosomes and pathogens [27][29][33][34]. In plants, a substantial number of autophagy receptors have also been identified, characterized and functionally analyzed. These plant autophagy receptors include Arabidopsis protein TSPO (outer membrane tryptophan-rich sensory protein), an ATG8-interacting heme-binding protein that targets the degradation of porphyrins through autophagy [35]. TSPO also targets the aquaporin PIP2;7 for degradation in the vacuole to reduce its levels to regulate water permeability under conditions of heat and drought stress [36][37]. ATG8-interacting 1 (ATI1) and 2 (ATI2) proteins are closely related autophagy receptors partially associated with the ER under normal conditions but mainly associated with a different type of spherical compartments under carbon starvation [38][39]. ATI1 is also found on bodies associated with plastids under carbon starvation and is involved in autophagy-dependent trafficking of plastid proteins to the vacuole [39]. The three related ATI3 autophagy receptors from Arabidopsis play a critical role in plant heat tolerance and resistance to the necrotrophic fungal pathogens at least in part through interaction with ER-localized UBAC2 proteins and mediating selective autophagy of specific unknown ER components [40]. Arabidopsis DSK2 acts as a selective autophagy receptor that targets BR1-EMS suppressor 1(BES1) to modulate brassinosteroid signaling and stress tolerance [41]. Arabidopsis ORM1 and 2 proteins mediate selective autophagy of pattern recognition receptor FLS2 to negatively regulate FLS2-mediated defense [42]. More recently, Arabidopsis cytosolic protein C53 has been identified as a receptor for selective autophagy of certain ER domains (ER-phagy) [43]. C53 is also an ER-phagy receptor in mammalian cells [44]. The proteasome subunit RPN10 is a selective autophagy receptor that mediates selective autophagy of the 26S proteasome when the proteasome is inhibited chemically [31]. RPN10 interacts with ATG8 through its UIM-like sequence and is increasingly associated with the proteasome when it is ubiquitinated [30].
While many selective autophagy receptors identified in plants are plant-specific, others are evolutionarily conserved and have homologs in nonplant organisms. Plant NBR1 is structurally related to human NBR1 and SQSTM1/p62 selective autophagy receptors that act in aggrephagy [14][45][46]. Since their first reports almost ten years ago [45,46], a number of studies have revealed an important role of NBR1 in plant responses to a broad spectrum of stress conditions including heat, salt, drought and oxidative stress [14][15][47]. More recently, new discoveries have been made in the understanding the roles and mode of action of NBR1 in the modulation of plant heat stress memory, plant-viral interaction and other physiological processes associated with plant stress responses.
Plant NBR1 is a structural homolog and functional hybrid of mammalian autophagy receptors NBR1 and p62, which differ in size but share similar domains and important features [45][46][48][49]. Both mammalian p62 and NBR1 proteins contain an N-terminal PB1 (Phox and Bem1p) domain followed by a ZZ-type zinc finger domain, an LC3-interacting region or LIR motif (also known as AIM motif in yeast) and a C-terminal UBA (ubiquitin-associated) domain [49]. As in many other selective autophagy receptors, the LIR motifs mediate direct interaction with ATG8, while the C-terminal UBA domain recognizes mono- and polyubiquitin. The PB1 domain of p62 mediates polymerization of p62, but the related human NBR1 protein lacks the most N-terminal basic charge cluster and, therefore, cannot polymerize through its PB1 domain [49]. However, human NBR1 can form heterodimers with p62 via its PB1 and can self-interacts via a coiled coil domain [49]. Mammalian p62 and NBR1 function as selective autophagy receptors for autophagic degradation of protein aggregates (aggrephagy), mitochondria (mitophagy), peroxisomes (pexophagy), endoplasmic reticulum (reticulophagy) and pathogens (xenophagy) [50]. p62 and NBR1 also have noncanonical role in signaling independent of autophagy [51][52][53][54]. For example, p62 promotes the expression of inflammatory genes via NF-κB through TRAF6 binding by its TRAF6-binding (TB) domain [53]. p62 can also activate mTORC1, which can upregulate c-Myc [55]. These functions are independent of the UBA or LC3-interacting region (LIR) domains of p62, which are required for its role as an autophagy receptor [53].
Even though NBR1 has a domain architecture similar to that of p62, it has a highly conserved globular domain characterized by the presence of 4 highly conserved tryptophan (W) residues that is absent in p62 [49]. Using this four tryptophan or FW domain to distinguish p62 and NBR1 homologs through the eukaryotic kingdom, it was discovered that only metazoans contain both p62 and NBR1 homologs, while most nonmetazonan organisms have a single NBR1 homolog [49]. Plant NBR1 homologs lack the coiled coil domain of mammalian NBR1 but have two C-terminal UBA domains [45]. In model plant Arabidopsis, there is a single gene encoding an NBR1 homolog. Like p62, Arabidopsis NBR1 can homo-oligomerizes through the N-terminal PB1 domain, indicating the plant NBR1 share some functional properties with p62 [45]. In addition, only the C-terminal UBA domain of the two UBA domains of Arabidopsis NBR1 binds ubiquitin. We have searched the sequenced genomes of representative plants along the evolutionary tree for genes encoding NBR1 homologs and identified 27 NBR1-encoding genes from 2 spore-bearing and 15 seed plants (Figure 1). As previously reported, some plants contain a single gene encoding NBR1. However, most plants including the spreading earthmoss (Physcomitrella patens), tomato, potato, rice, maize, purple false brome (Brachypodium distachyon) and soybean contain two genes encoding NBR1 proteins (Figure 1). Maize, soybean and purple false brome are known to be polyploid plants that have gone genome duplications during their evolutionary history [56–58], which could account for the presence of more than one NBR1 genes in their genomes. To analyze the evolutionary relationship of the conserved protein family, we performed phylogenetic analysis of NBR1 homologs from 17 plant and 4 animal species. As shown in Figure 1, there are three major clades in the phylogenetic tree. All the NBR1 proteins from the animals clustered in one clade, which intriguingly also include the NPR1 from Aquilegia coerulea, while those from plants clustered in two separate clades (Figure 1). These results indicate that the topology of phylogenetic tree for NBR1 homologs from animals and plants is in general agreement with the evolutionary tree of the organisms. However, there is no clear separation of clustering of NBR1 homologs between spore-bearing and seed plants and likewise, NBR1 proteins from monocot and dicot plants do not clustered separately in the clades of seed plants (Figure 1). Furthermore, while the twin NBR1 homologs from some plants such as soybean and P. patens clustered together, other twin NBR1 homologs from the same plant species including tomato, potato, rice, maize and purple false brome do not group together in the tree (Figure 1), indicating significant sequence divergence between these twin homologs. The evolutionary significance of the sequence variation of the NBR1 homologs is unclear but could reflect potential functional diversification of the two NBR1 homologs in these plants.
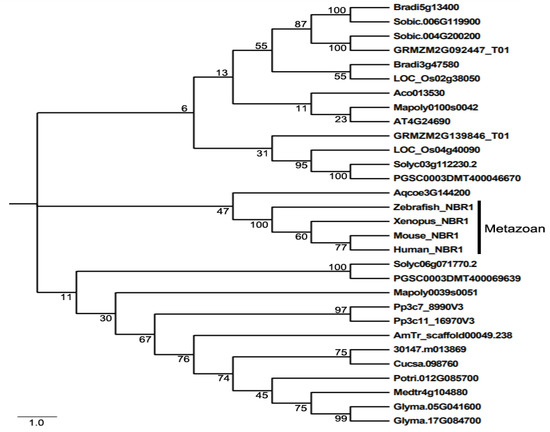
Figure 1. The phylogenetic relationship of NBR1 homologs from plants and metazoans. The tree was inferred using the neighbor-joining method. Phylogenetic analyses were conducted in MEGA5. Bootstrap values from 1000 replicates were used to assess the robustness of the tree. NBR1 homologs in the phylogenetic analysis include those from Homo sapiens (Human NBR1), Mus musculus (Mouse NBR1), Danio rerio (Zebrafish NBR1), Xenopus tropicalis (Xenopus NBR1), Marchantia polymorpha (Mapoly0100s0042 and Mapoly0039s0051), Physcomitrella patens (Pp3c7_8990V3 and Pp3c11_16970V3), Amborella trichopoda (AmTr scaffold00049.238), Ananas comosus(Aco013530), Brachypodium distachyon (Bradi3g47580 and Bradi5g13400), Oryza sativa (LOC_Os02g38050 and LOC_Os04g40090), Sorghum bicolor (Sobic.004G200200 and Sobic.006G119900), Zea mays (GRMZM2G139846_T01 and GRMZM2G092447_T01), Aquilegia coerulea (Aqcoe3G144200), Solanum lycopersicum (Solyc03g112230.2 and Solyc06g071770.2), Solanum tuberosum (PGSC0003DMT400069639 and PGSC0003DMT400046670), Populus trichocarpa (Potri.012G085700), Ricinus communis (30147.m013869), Arabidopsis thaliana (AT4G24690), Cucumis sativus (Cucsa.098760), Glycine max (Glyma.05G041600 and Glyma.17G084700), Medicago truncatula (Medtr4g104880).
A number of groups have reported functional genetic analysis of plant NBR1 through characterization of nbr1 mutants or transgenic silencing lines [14][15][46][47][56][57][58][59]. Arabidopsis nbr1 knockout mutants grow and develop normally under normal growth conditions and is not essential for general autophagy, or for the selective clearance of peroxisomes, mitochondria, or the ER [14][57][59]. Plant NBR1 is also dispensable in age- and darkness-induced senescence but may modulates senescence under certain conditions such as short-day growth condition [14][56]. The Arabidopsis nbr1 mutants are also normal in resistance to a necrotrophic pathogen [14]. However, the nbr1 mutants are compromised in plant tolerance to heat, oxidative, salt, and drought stresses [14][47]. The role of NBR1 in plant abiotic stress tolerance is dependent on its interaction with ATG8. In Arabidopsis, NBR1 also reduces growth of bacterial pathogen Pseudomonas syringae by suppressing the establishment of an aqueous extracellular space (‘water-soaking’) [58]. Therefore, the nbr1 mutants display some but not all of the phenotypes of autophagy-deficient mutants. As will be discussed below, more recent studies have revealed specific cargo proteins of plant NBR1 and their roles in the modulation of plant heat stress memory, plant-viral interaction, senescence, reactive oxygen species-induced pexophagy and nutrient responses.
As a structural and functional homolog of mammalian p62 and NBR1, plant NBR1 also acts as selective receptor for aggrephagy that targets misfolded proteins and protein aggregates that are generated under a variety of stress conditions. The conserved role of plant NBR1 in selective autophagy likely underlies its critical roles in plant responses to a broad spectrum of biotic and abiotic stresses including heat, drought, salt and oxidative stresses. Plant NBR1 also target specific protein cargoes including heat induced protein chaperones, which paradoxically reduces heat stress memory and compromises tolerance to subsequent heat stress, probably as a mechanism to expedite the recovery of plant growth after the cessation of a heat stress. NBR1 is also increasingly recognized as a critical player in plant antiviral immunity by directly targeting specific viral proteins for degradation in the vacuole. This role of plant NBR1 in plant antiviral defense and counter-defense mechanism through exploitation of plant autophagy by viral pathogens can open new frontiers in the study of the dynamic and complex interactions between plants and microbial pathogens. Through targeting specific target proteins, NBR1-mediated selective autophagy also participates in the modulation of other important processes in plants including stress-induced pexophagy, S nutrient responses and ABA signaling. Despite the progresses, our understanding of NBR1-mediated selective autophagy is still at its infancy. The number of identified cargo proteins for plant NBR1 is still very limited and, as a result, the underlying mechanisms for the broad biological functions of plant NBR1 are still not well understood. Currently there is no comprehensive knowledge about the pathways that regulate the protein levels and activity of plant NBR1 beyond the established fact that the selective autophagy receptor is itself subjected to degradation in the vacuole during autophagy. Animal NBR1 proteins also have novel roles in signaling independent of autophagy and it remains to be determined whether plant NBR1 has similar regulatory roles in signaling in a manner independent of autophagy. A better knowledge about the broad and complex roles of NBR1 will provide new important insights into the molecular basis of plant responses to biotic and abiotic stresses.