1000/1000
Hot
Most Recent

Chemo and radiation therapies are the most commonly used therapies for cancer, but they can induce DNA damage, resulting in the apoptosis of host cells. DNA double-stranded breaks (DSBs) are the most lethal form of DNA damage in cells, which are constantly caused by a wide variety of genotoxic agents, both environmentally and endogenously. To maintain genomic integrity, eukaryotic organisms have developed a complex mechanism for the repair of DNA damage. Researches reported that many cellular long noncoding RNAs (lncRNAs) were involved in the response of DNA damage. The roles of lncRNAs in DNA damage response can be regulated by the dynamic modification of N6-adenosine methylation (m6A). The cellular accumulation of DNA damage can result in various diseases, including cancers. Additionally, lncRNAs also play roles in controlling the gene expression and regulation of autophagy, which are indirectly involved with individual development. The dysregulation of these functions can facilitate human tumorigenesis. In this review, we summarized the origin and overview function of lncRNAs and highlighted the roles of lncRNAs involved in the repair of DNA damage.
DNA damage is constantly caused by various endogenous and exogenous factors, such as ionizing radiation, ultra-violet, reactive oxygen species (ROS), and genotoxic drugs [1][2]. It is generally accepted that DNA damage is a potential threat to human health. Human have evolved intricate mechanisms for the repair of DNA damage to sustain genome stability, and homologous recombination (HR) and nonhomologous end joining (NHEJ), as two major DSBs repair pathways, have been ubiquitously applied in cells [3][4]. If living organisms fail to accurately repair the damaged DNA in cells, the accumulation of DNA damage will lead to serious consequences and, eventually, the occurrence of cancers in the body. So, genomic integrity is essential for organism survival and for the inheritance of traits to offspring. Long noncoding RNAs (LncRNAs) are an important class of RNA transcripts, with over 200 nucleotides in length, which resemble protein-coding genes but lack the ability for translation into proteins in general [5]. Hangauer et al. [6] reported that over 10,000 lncRNA transcripts could be produced from the human genome, and some lncRNAs were reported to play regulatory roles in various biological processes, ranging from the innate immune response, cell cycle control, pluripotency, and differentiation to disease [7][8][9]. Moreover, recent evidence showed that some lncRNAs such as NORAD and GUARDIN could directly participate in the repair of DNA damage [9][10][11].
Different classes of lncRNAs were transcribed from several DNA elements, such as enhancers, promoters, and intergenic regions, in eukaryotic genomes [12]. Iyer et al. (2015) [13] reported that over 50,000 lncRNAs (designated MiTranscriptome lncRNAs) could be generated in the human transcriptome from various tumors, normal tissues, and cell lines based on The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/). To date, 268,848 lncRNAs have been collected in the database of human lncRNAs (https://bigd.big.ac.cn/lncbook/index), which is far greater than the number of protein-coding mRNAs (~20,000) in the genome. Unlike protein-coding mRNAs, lncRNAs exhibit functional uniqueness by participating in and modulating various cellular processes, including histone modification, DNA methylation, cellular transcription, the inflammatory response, antiviral immunity, and repair of DNA damage [14][15][16][17]. Additionally, some lncRNAs also function as diagnostic markers and/or possible therapeutic targets. Therefore, the understanding of biogenesis and the biological functions of lncRNAs is helpful for disclosing their functional significance.
According to the diversity of noncoding RNAs, they can be divided into two main types: structural noncoding RNAs and regulatory noncoding RNAs [8]. Structural noncoding RNAs comprise of rRNAs and tRNAs, and regulatory noncoding RNAs are further divided into three classes: small, medium, and long noncoding RNAs (Figure 1A) [18][19]. The biogenesis of lncRNAs is cell type- and stage-specific, which is under the control of cell type- and stage-specific stimuli. Different classes of lncRNAs were reported to be transcribed from different DNA elements, such as enhancers, promoters, and intergenic regions, in eukaryotic genomes (Figure 1B). As we know, promoters and enhancers are essential DNA elements in the control of gene expression networks. Some short-lived medium-length lncRNAs can be transcribed from promoter upstream regions and enhancers by RNA polymerase II (Pol II), and some lncRNAs can be bidirectionally transcribed from enhancers by Pol II [20][21]. Additionally, some lncRNAs are transcribed by Pol II from intergenic regions between two genes and represent the best-studied subclass of lncRNAs.
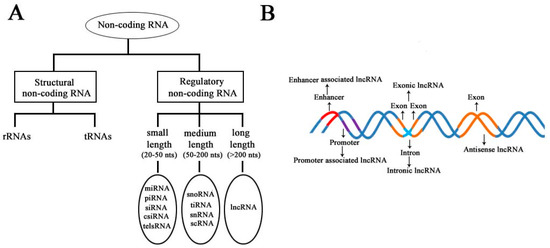
Figure 1. Schematic diagram to illustrate the diversity of long noncoding RNAs (lncRNAs) in the mammalian genome. (A) Classification of noncoding RNAs according to their size and function. (B) Overview of the biogenesis of various lncRNAs through different mechanisms. Different colors indicate different regions of DNA elements in the mammalian genome.
Most annotated lncRNAs contain multiple exons and have typical mRNA-like features, with a 5′ m7G cap and a 3′ poly(A) tail. These similarities existing between lncRNAs and mRNAs provide the possibility that mature lncRNAs may behave similarly to mRNAs in cells. In fact, this is not the truth. Due to a lacking of robust protein-coding potential, lncRNAs are less evolutionarily conserved and less abundant. They exhibit more tissue-specific expression and greater nuclear localization patterns. Additionally, a significant difference was found among different lncRNAs varying in their sizes. In the database of lncRNA (http://lncrnamap.mbc.nctu.edu.tw/php), the statistics of the lncRNA classes show that there are 23,879 lncRNAs with length <1000 nt, 4985 lncRNAs with lengths ranging from 1000 to 2000 nt, 1943 lncRNAs with lengths ranging from 2000 to 3000 nt, and 12 lncRNAs with lengths ranging from 9000 to 10,000 nt). Moreover, some lncRNAs were found to be involved in the DNA damage response, which are summarized in Table 1.
Table 1. Different long noncoding RNA (lncRNA) involvement with DNA damage.
| LncRNAs | Accession Number |
Functions | Length (nts) | Genome | Refs. |
|---|---|---|---|---|---|
| NORAD | NR_027451.1 | Critical for genome stability | 5378 | Human | [10] |
| GUARDIN | NR_132738.1 | Critical for genome stability | 1003 | Human | [11] |
| LCPAT1 | NM_018715.4 | Involvement with DNA damage | 4040 | Human | [17] |
| LINC00261 | NR_001558.3 | Activation of DDR | 4924 | Human | [21] |
| Meg3 | NR_046473.1 | Regulation of DNA damage response | 9701 | Human | [22] |
| DNM3OS | NR_038397.2 | Regulation of DNA damage response | 7957 | Human | [23] |
| LINP1 | NR_138480.1 | facilitating DNA damage repair | 838 | Human | [24] |
| DINO | NR_144384.1 | Efficient activation of p53 target genes | 951 | Human | [25] |
| TODRA | NR_040058.1 | Promoting RAD51-dependent DSB repair | 1156 | Human | [26] |
| DDSR1 | KT318134.1 | Modulating DNA repair by HR | 1616 | Human | [27] |
| JADE | KC469579.1 | Functional linking with histone H4 acetylation | 1721 | Human | [28] |
| NEAT1 | MK562403.1 | A common mediator for inflammasome stimuli | 2713 | Human | [29] |
| ROR | HG975412.1 | A p53 repressor in response to DNA damage | 2591 | Human | [30] |
Note. DDR and HR are the abbreviations for DNA damage response and homologous recombination. DSB: double-stranded break. Refs is the abbreviations for references.
N6-adenosine methylation (m6A) is the most common internal modification in mRNA and long noncoding RNA, and it is also a dynamic reversible modification with implications in fine-tuning the cellular metabolism. It is modulated by m6A regulators, including “writers” (methyltransferases), “readers” (signal transducers), and “erasers” (demethylases) [31]. To date, this modification has been identified in various organisms, including yeasts, plants, flies, mammals, and some viruses, and more than 12,000 m6A sites in the transcripts of ∼7000 protein-coding genes and ∼300 noncoding genes have been characterized in human cells [32]. Furthermore, the majority of m6A was found within the conserved RRACH motif (R = G/A and H = A/C/U) in mRNAs. Meanwhile, many lncRNAs could be modified by m6A, which can control many aspects of gene expression and cellular biology at both the transcriptional and post-transcriptional levels [33][34][35].
However, the aberrant expression and dysregulation of lncRNA is strongly linked to tumorigenesis, metastasis, and the tumor stage [36][37]. For example, MEG3 and NBAT1 have been confirmed to play an important role in the formation of pathogenicity of gliomas [38][39]. Moreover, MALAT1 is highly expressed in the nucleus, and it has been confirmed to play a suppressive role in the formation of gliomas by downregulating MMP2 and devitalizing ERK/MAPK signaling [40]. The m6A modification has been confirmed to play functional roles in RNA splicing, nuclear export, and decay [41]. For example, the MALAT1 with m6A modification could regulate the interaction between RNAs and some special binding proteins and, also, affect its localization and activity in the nucleus [42]. Now, the m6A modification has been identified as the most abundant modification in mRNA and noncoding RNA (ncRNA). Accumulating studies have focused on the role of lncRNAs regulated by m6A modification in cancer progression, and it was used to demonstrate the mechanisms by which m6A participates in the biology of cancers.
As is well-known, DNA damage is closely involved with the occurrence and development of cancers, and many lncRNAs were involved in the repair of DNA damage [43][44]. To further examine whether the expression of lncRNAs is regulated by m6A, an analysis of RT-qPCR was performed to detect the expressions of some lncRNAs related to DNA damage. For the purpose, some lncRNAs from siControl- and siWTAP-transfected HCC cell lines (SMCC7721) were selected for the analysis. The results showed that the expressions of ROR, LINP1, TERRA, and DNM3OS were significantly increased by over two-fold compared with the control group (Figure 2, unpublished data). Additionally, the expressions of DDSR1, SNHG5, LCPAT1, NORAD, and ANRIL also showed a 1.5-fold increase compared with the control group, and the statistical analysis further showed that there were significant differences between siControl and siWTAP (Figure 2). As far as the rest of the lncRNAs (Figure 2) were concerned, no obvious changes in the expression levels were observed compared with the control group. Therefore, we think that the expressions of some lncRNAs related to DNA damage could be regulated by m6A, indicating that the modification may play an important role in the regulation of the DNA damage response. However, abnormal regulation may directly promote tumorigenesis.
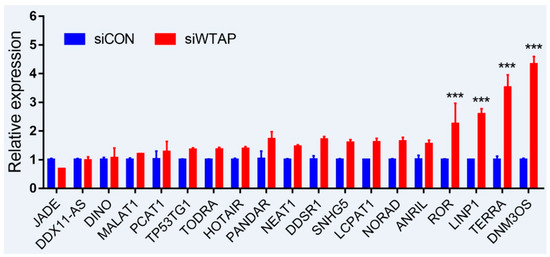
Figure 2. Expression analysis of lncRNAs through RT-qPCR from the siControl- and siWTAP-transfected HCC cell lines (SMCC7721). The error bar represents the standard deviation of each mean value (mean ± SD; n = 3). Asterisks indicate significant differences compared with the controls. *** represents significant difference compared with control group.