The endospores (spores) of many Bacillus cereus sensu lato species are decorated with multiple hair/pilus-like appendages. Although they have been observed for more than 50 years, all efforts to characterize these fibers in detail have failed until now, largely due to their extraordinary resilience to proteolytic digestion and chemical solubilization. A recent structural analysis of B. cereus endospore appendages (Enas) using cryo-electron microscopy has revealed the structure of two distinct fiber morphologies: the longer and more abundant “Staggered-type” (S-Ena) and the shorter “Ladder-like” type (L-Ena), which further enabled the identification of the genes encoding the S-Ena. Ena homologs are widely and uniquely distributed among B. cereus sensu lato species, suggesting that appendages play important functional roles in these species. The discovery of ena genes is expected to facilitate functional studies involving Ena-depleted mutant spores to explore the role of Enas in the interaction between spores and their environment.
- endospore
- spore
- appendage
- Bacillus cereus
- Ena
1. Endospore Appendages
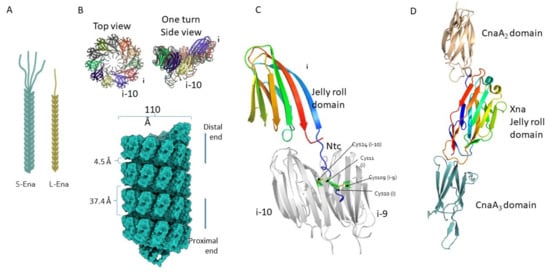
2. Architecture of Ena Fibers
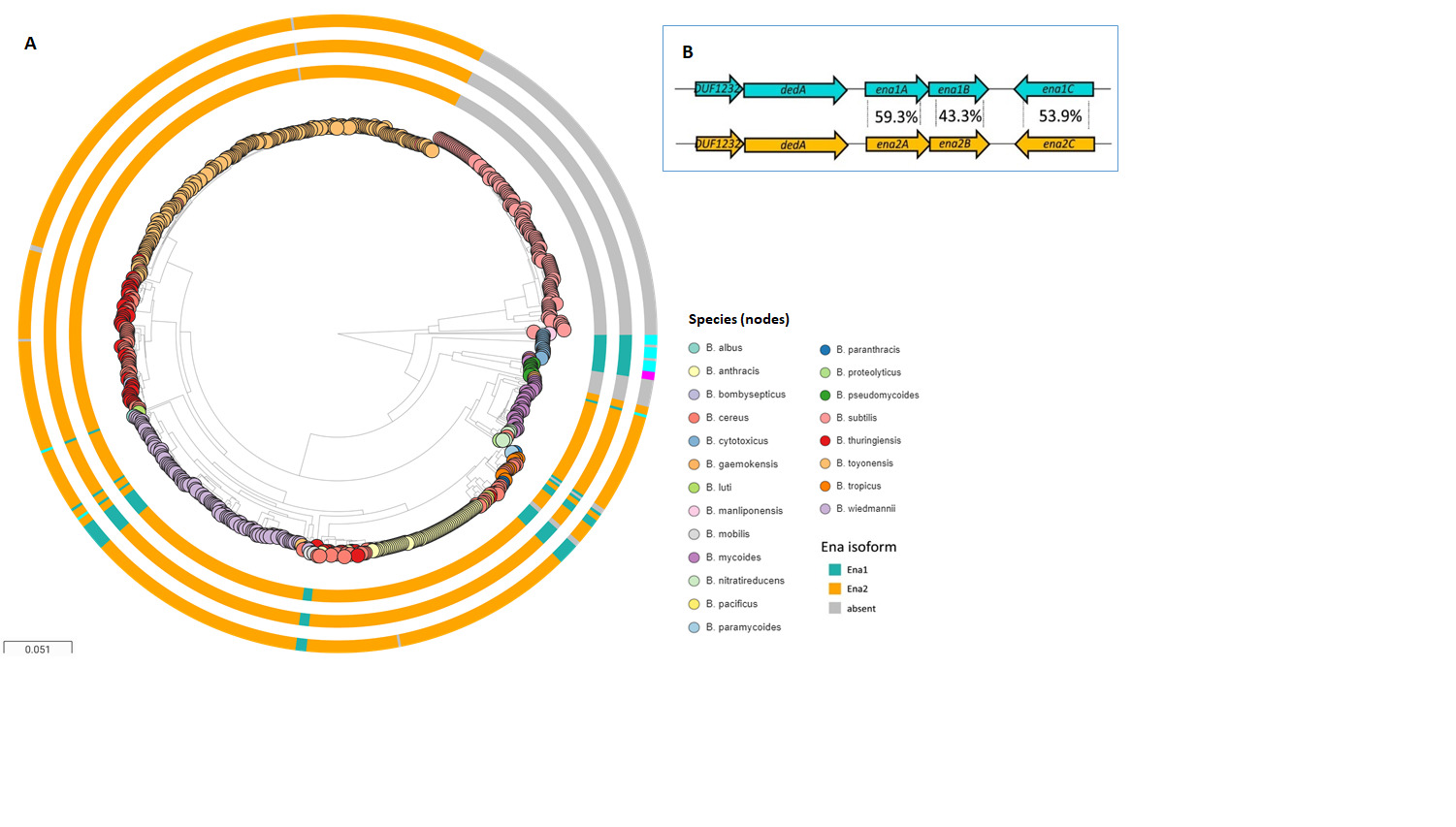
3. Ena-Encoding Genes
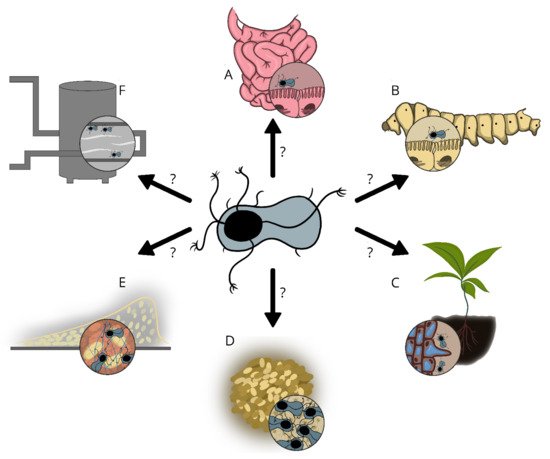
4. Potential Functions of Enas
5. Concluding Remarks
The Enas are suggested to play a role in adhesion to biotic and/or abiotic surfaces, but sufficient experimental data are lacking. The recent discovery of the genes encoding the predominant type of Enas (S-Ena) in B. cereus s.l. spp.[7], is expected to facilitate comparative studies involving Ena depleted mutant spores and wild type spores. Such studies will give important insights into the function of these extraordinary fibers in spore adherence to biotic and abiotic surfaces, biofilm formation, spore aggregation, germination, virulence, and other phenomena that would have important implications in the biology of these species. Importantly, knowledge of the potential function of Enas in spore adhesion would also allow the design of more effective strategies to prevent spore binding when harmful or promote binding when beneficial.
This entry is adapted from the peer-reviewed paper 10.3390/ijms222212367
References
- Rode, L.J.; Crawford, M.A.; Williams, M.G. Clostridium spores with ribbon-like appendages. J. Bacteriol. 1967, 93, 1160–1173.
- Hachisuka, Y.; Kuno, T. Filamentous appendages of Bacillus cereus spores. Jpn. J. Microbiol. 1976, 20, 555–558.
- Ankolekar, C.; Labbé, R.G. Physical characteristics of spores of food-associated isolates of the Bacillus cereus group. Appl. Environ. Microbiol. 2010, 76, 982–984.
- Hachisuka, Y.; Kozuka, S. A new test of differentiation of Bacillus cereus and Bacillus anthracis based on the existence of spore appendages. Microbiol. Immunol. 1981, 25, 1201–1207.
- Hachisuka, Y.; Kozuka, S.; Tsujikawa, M. Exosporia and appendages of spores of Bacillus species. Microbiol. Immunol. 1984, 28, 619–624.
- Tauveron, G.; Slomianny, C.; Henry, C.; Faille, C. Variability among Bacillus cereus strains in spore surface properties and influence on their ability to contaminate food surface equipment. Int. J. Food Microbiol. 2006, 110, 254–262.
- Pradhan, B.; Liedtke, J.; Sleutel, M.; Lindbäck, T.; Zegeye, E.D.; O’Sullivan, K.; Llarena, A.; Brynildsrud, O.; Aspholm, M.; Remaut, H. Endospore Appendages: A novel pilus superfamily from the endospores of pathogenic Bacilli. EMBO J. 2021, 40, e106887.
- Plomp, M.; Leighton, T.J.; Wheeler, K.E.; Malkin, A.J. Architecture and high-resolution structure of Bacillus thuringiensis and Bacillus cereus spore coat surfaces. Langmuir 2005, 21, 7892–7898.
- Budzik, J.M.; Poor, C.B.; Faull, K.F.; Whitelegge, J.P.; He, C.; Schneewind, O. Intramolecular amide bonds stabilize pili on the surface of bacilli. Proc. Natl. Acad. Sci. USA 2009, 106, 19992–19997.
- Lukaszczyk, M.; Pradhan, B.; Remaut, H. The biosynthesis and structures of bacterial pili. In Bacterial Cell Walls and Membranes; Subcellular Biochemistry; Springer: Cham, Switzerland, 2019; Volume 92.
- Walker, J.R.; Gnanam, A.J.; Blinkova, A.L.; Hermandson, M.J.; Karymov, M.A.; Lyubchenko, Y.L.; Graves, P.R.; Haystead, T.A.; Linse, K.D. Clostridium taeniosporum spore ribbon-like appendage structure, composition and genes. Mol. Microbiol. 2007, 63, 629–643.
- Kozuka, S.; Tochikubo, K. Properties and origin of filamentous appendages on spores of Bacillus cereus. Microbiol. Immunol. 1985, 29, 21–37.
- Stalheim, T.; Granum, P.E. Characterization of spore appendages from Bacillus cereus strains. J. Appl. Microbiol. 2001, 91, 839–845.
- Kozuka, S. Fragmentation and solubilization of filamentous appendages of Bacillus cereus spores. Nippon. Saikingaku Zasshi. Jpn. J. Bacteriol. 1993, 8, 541–550.
- Callaway, E. “It opens up a whole new universe”: Revolutionary microscopy technique sees individual atoms for first time. Nature 2020, 582, 156–157.
- Richardson, J.S. The anatomy and taxonomy of protein structure. Adv. Protein Chem. 1981, 34, 167–339.
- Kailas, L.; Terry, C.; Abbott, N.; Taylor, R.; Mullin, N.; Tzokov, S.B.; Todd, S.J.; Wallace, B.A.; Hobbs, J.K.; Moir, A.; et al. Surface architecture of endospores of the Bacillus cereus/anthracis/ thuringiensis family at the subnanometer scale. Proc. Natl. Acad. Sci. USA 2011, 108, 16014–16019.
- Hae, J.K.; Paterson, N.G.; Gaspar, A.H.; Hung, T.T.; Baker, E.N. The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds. Proc. Natl. Acad. Sci. USA 2009, 106, 16967–16971.
- Shaik, M.M.; Maccagni, A.; Tourcier, G.; Di Guilmi, A.M.; Dessen, A. Structural basis of pilus anchoring by the ancillary pilin RrgC of Streptococcus pneumoniae. J. Biol. Chem. 2014, 289, 16988–16997.
- Mishra, A.; Devarajan, B.; Reardon, M.E.; Dwivedi, P.; Krishnan, V.; Cisar, J.O.; Das, A.; Narayana, S.V.L.; Ton-That, H. Two autonomous structural modules in the fimbrial shaft adhesin FimA mediate Actinomyces interactions with streptococci and host cells during oral biofilm development. Mol. Microbiol. 2011, 81, 1205–1220.
- Persson, K.; Esberg, A.; Claesson, R.; Strömberg, N. The pilin protein FimP from Actinomyces oris: Crystal structure and sequence analyses. PLoS ONE 2012, 7, e48364.
- Deivanayagam, C.C.S.; Rich, R.L.; Carson, M.; Owens, R.T.; Danthuluri, S.; Bice, T.; Höök, M.; Narayana, S.V.L. Novel fold and assembly of the repetitive B region of the Staphylococcus aureus collagen-binding surface protein. Structure 2000, 8, 67–78.
- Hartman, R.; Eilers, B.J.; Bollschweiler, D.; Munson-McGee, J.H.; Engelhardt, H.; Young, M.J.; Lawrence, C.M. The molecular mechanism of cellular attachment for an Archaeal Virus. Structure 2019, 27, 1634–1646.
- Khare, B.; Narayana, S.V.L. Pilus biogenesis of Gram-positive bacteria: Roles of sortases and implications for assembly. Protein Sci. 2017, 26, 1458–1473.
- Kang, H.J.; Baker, E.N. Intramolecular isopeptide bonds give thermodynamic and proteolytic stability to the major pilin protein of Streptococcus pyogenes. J. Biol. Chem. 2009, 284, 20729–20737.
- Katz, L.; Griswold, T.; Morrison, S.; Caravas, J.; Zhang, S.; Bakker, H.; Deng, X.; Carleton, H. Mashtree: A rapid comparison of whole genome sequence files. J. Open Source Softw. 2019, 4, 1762.
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132.
- Argimón, S.; Abudahab, K.; Goater, R.J.E.; Fedosejev, A.; Bhai, J.; Glasner, C.; Feil, E.J.; Holden, M.T.G.; Yeats, C.A.; Grundmann, H.; et al. Microreact: Visualizing and sharing data for genomic epidemiology and phylogeography. Microb. Genom. 2016, 2, e000093.
- Swick, M.C.; Koehler, T.M.; Driks, A. Surviving between hosts: Sporulation and transmission. Microbiol. Spectr. 2016, 4, 26.
- Petrova, O.E.; Sauer, K. Sticky situations: Key components that control bacterial surface attachment. J. Bacteriol. 2012, 194, 2413–2425.
- Andersson, A.; Granum, P.E.; Rönner, U. The adhesion of Bacillus cereus spores to epithelial cells might be an additional virulence mechanism. Int. J. Food Microbiol. 1998, 39, 93–99.
- Ramarao, N.; Lereclus, D. Adhesion and cytotoxicity of Bacillus cereus and Bacillus thuringiensis to epithelial cells are FlhA and PlcR dependent, respectively. Microbes Infect. 2006, 8, 1483–1491.
- Kline, K.A.; Fälker, S.; Dahlberg, S.; Normark, S.; Henriques-Normark, B. Bacterial adhesins in host-microbe interactions. Cell Host Microbe 2009, 5, 580–592.
- Ton-That, H.; Schneewind, O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 2003, 50, 1429–1438.
- Izoré, T.; Contreras-Martel, C.; El Mortaji, L.; Manzano, C.; Terrasse, R.; Vernet, T.; Di Guilmi, A.M.; Dessen, A. Structural basis of host cell recognition by the pilus adhesin from Streptococcus pneumoniae. Structure 2010, 18, 106–115.
- Wu, C.; Mishra, A.; Yang, J.; Cisar, J.O.; Das, A.; Ton-That, H. Dual function of a tip fimbrillin of Actinomyces in fimbrial assembly and receptor binding. J. Bacteriol. 2011, 193, 3197–3206.

