Cold-loving microorganisms of all three domains of life have unique and special abilities that allow them to live in harsh environments. They have acquired structural and molecular mechanisms of adaptation to the cold that include the production of anti-freeze proteins, carbohydrate-based extracellular polymeric substances and lipids which serve as cryo- and osmoprotectants by maintaining the fluidity of their membranes. They also produce a wide diversity of pigmented molecules to obtain energy, carry out photosynthesis, increase their resistance to stress and provide them with ultraviolet light protection. Recently developed analytical techniques have been applied as high-throughoutput technologies for function discovery and for reconstructing functional networks in psychrophiles. Among them, omics deserve special mention, such as genomics, transcriptomics, proteomics, glycomics, lipidomics and metabolomics. These techniques have allowed the identification of microorganisms and the study of their biogeochemical activities. They have also made it possible to infer their metabolic capacities and identify the biomolecules that are parts of their structures or that they secrete into the environment, which can be useful in various fields of biotechnology.
- biomolecules
- microorganisms
- proteins
- metabolites
- pigments
- antibiotics
- antifreeze molecules
- lipids
- omics
- metabolic routes
1. Introduction
The identification and characterization of new biomolecules from natural resources is a major challenge for today’s society. Therefore, new strategies and next-generation approaches are needed. Frozen environments constitute an ecological niche inhabited by microorganisms with very remarkable characteristics that produce specific biomolecules. Nowadays, the cryosphere covers about one-fifth of the surface of the Earth [1]. This includes the polar ice sheets, sea ice, ice caves and mountain glaciers ( Figure 1 ). The discovery of cold-tolerant microorganisms in permanently frozen environments has broadened the known range of environmental conditions which support microbial life. Most of the biomass of the cryosphere is made up of microorganisms. There are various classifications for cold-loving microorganisms, but the most used is that of Morita [2], which differentiates between psychrophiles and psychrotolerants or psychrotropes. As this review is more focused on biomolecules than on microorganisms, we will refer to all of them generically as psychrophiles [3].
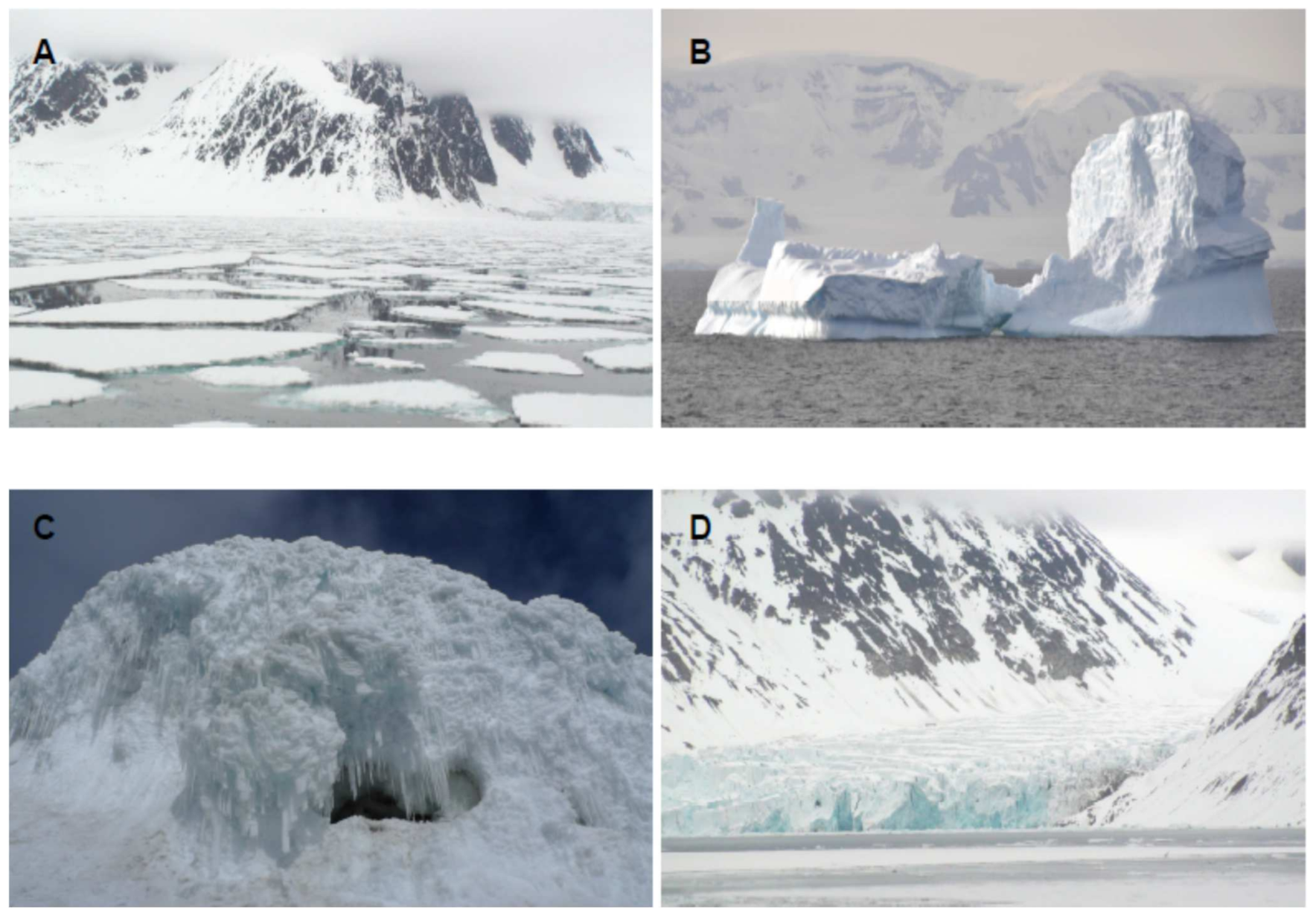 Figure 1. Representative environments inhabited by cold loving microorganisms. (A) Sea ice. (B) Iceberg. (C) Ice cave. (D) Glacier.
Figure 1. Representative environments inhabited by cold loving microorganisms. (A) Sea ice. (B) Iceberg. (C) Ice cave. (D) Glacier.
Microbial activity at cold temperatures is restricted to small amounts of unfrozen water inside the permafrost soil or the ice, and to seawater. Life thrives in these environments with an extraordinary microbial diversity of bacteria, archaea, fungi (in particular yeasts) and microalgae. The freezing process represents an important threat for the survival of life, as growing ice crystals can break cells and disrupt their membranes. Temperatures below 0 °C slow down cellular reaction rates by altering the functionality of the molecular building blocks. However, sometimes cells do not only tolerate this extreme environment, but require it to live. It is well documented that the temperature ranges of microbial habitats play a major role in the selection and adaptation of the resident microorganisms, and hence in microbial diversification [4]. Cold-adapted microorganisms possess molecules (lipids and proteins) in their cellular membranes which provide a flexible interface with the environment for the continued uptake of nutrients and the release of by-products. In this way, they maintain homeostasis and biochemical catalysis at low temperatures.
In recent years, a plethora of new techniques, notably omics, have revolutionized the study of environmental microorganisms. Omics include genomics, transcriptomics, proteomics, glycomics, lipidomics and metabolomics. These techniques have made it possible to discover a microscopic frozen world populated by microorganisms of a very diverse nature, capable of adapting to an extreme environment. These techniques have allowed us to know how these organisms live, and what types of adaptations to the environment allow them to survive.
2. Omics in Cold-Loving Microorganisms
It is true that the vast majority of polar microorganisms cannot be cultured or isolated, and new strategies and next generation approaches are needed to increase the chances of drug discovery (Figure 5). Omics comprise several and very different techniques, such as genomics, transcriptomics, proteomics, glycomics, lipidomics and metabolomics [5].
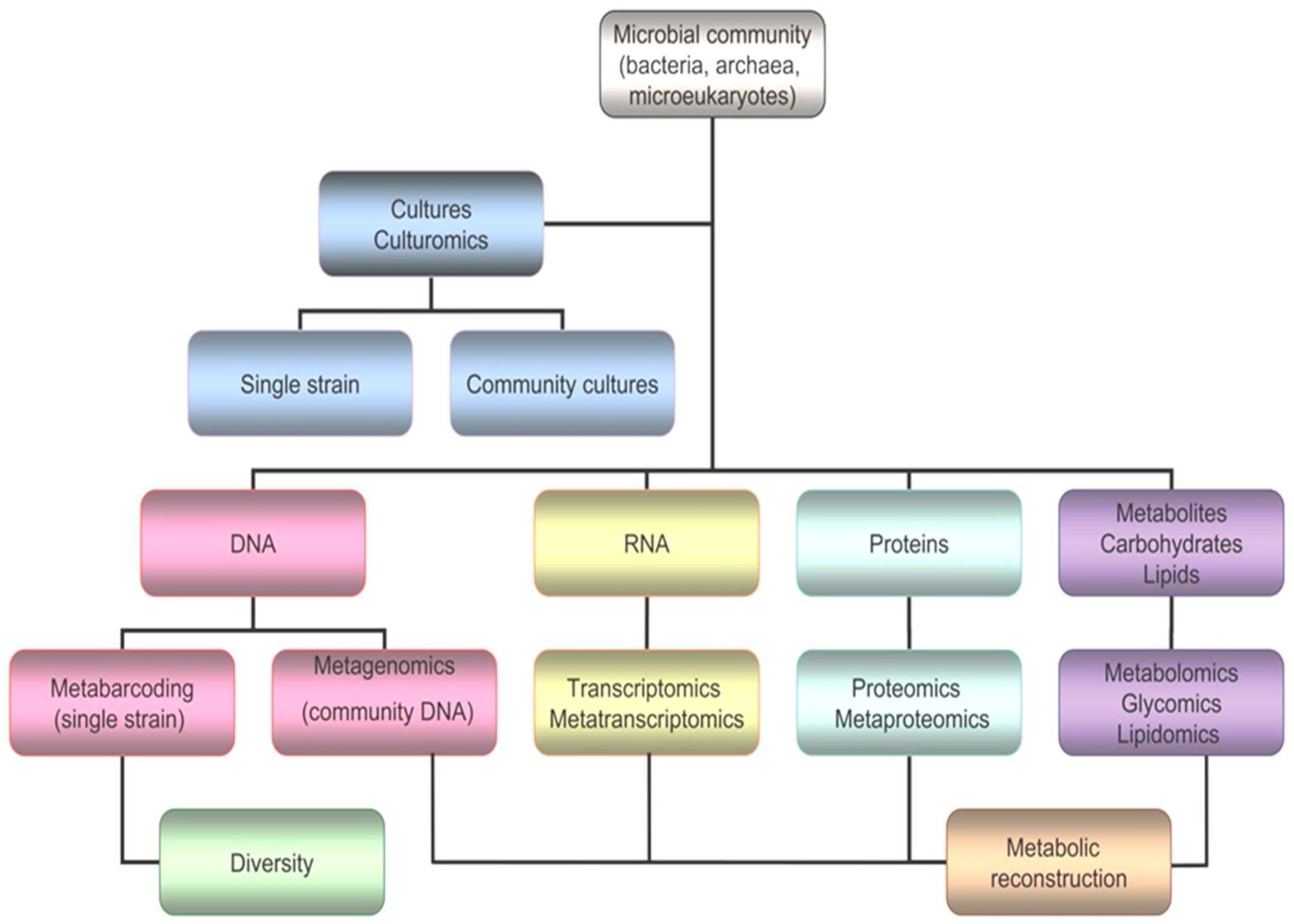
Figure 5. Workflow for omics, including genomics, transcriptomics, proteomics, glycomics, lipidomics and metabolomics.
2.1. Genomics
2.2. Transcriptomics and Metatranscriptomics
2.3. Proteomics and Metaproteomics
2.4. Glycomics and Lipidomics
2.5. Metabolomics
3. Conclusions and Perspectives
This entry is adapted from the peer-reviewed paper 10.3390/biom11081155
References
- Boetius, A.; Anesio, A.M.; Deming, J.W.; Mikucki, J.A.; Rapp, J. Microbial ecology of the cryosphere: Sea ice and glacial habitats. Nat. Rev. Genet. 2015, 13, 677–690.
- Craig, L.; Moyer, R.; Collins, R.; Morita, R.Y. Psychrophiles and Psychrotrophs. Reference Module in Life Sciences; Elsevier: Amsterdam, The Netherlands, 2017.
- Priscu, J.C.; Christner, B.C. Earth’s icy biosphere. In Microbial Diversity and Bioprospecting; Bull, A., Ed.; ASM Press: Washington, DC, USA, 2004; pp. 130–145.
- Alcazar, A.; Garcia-Descalzo, L.; Cid, C. Microbial evolution and adaptation in icy worlds. Astrobiology: Physical Origin, Biological Evolution and Spatial Distribution; Springer: New York, NY, USA, 2010; pp. 81–95.
- Maghembe, R.; Damian, D.; Makaranga, A.; Nyandoro, S.S.; Lyantagaye, S.L.; Kusari, S.; Hatti-Kaul, R. Omics for Bioprospecting and Drug Discovery from Bacteria and Microalgae. Antibiotics 2020, 9, 229.
- Raymond-Bouchard, I.; Whyte, L.G. From Transcriptomes to Metatranscriptomes: Cold Adaptation and Active Metabolisms of Psychrophiles from Cold Environments. In Psychrophiles: From Biodiversity to Biotechnology; Margesin, R., Ed.; Springer: Cham, Germany, 2017.
- Heueis, N.; Vockenhuber, M.-P.; Suess, B. Small non-coding RNAs in streptomycetes. RNA Biol. 2014, 11, 464–469.
- Martinez-Alonso, E.; Pena-Perez, S.; Serrano, S.; Garcia-Lopez, E.; Alcazar, A.; Cid, C. Taxonomic and functional characterization of a microbial community from a volcanic englacial ecosystem in Deception Island, Antarctica. Sci. Rep. 2019, 9, 1–14.
- García-Descalzo, L.; Alcazar, A.; Baquero, F.; Cid, C. Identification of in vivo HSP90-interacting proteins reveals modularity of HSP90 complexes is dependent on the environment in psychrophilic bacteria. Cell Stress Chaperon. 2010, 16, 203–218.
- Adibekian, A.; Stallforth, P.; Hecht, M.-L.; Werz, D.B.; Gagneux, P.; Seeberger, P.H. Comparative bioinformatics analysis of the mammalian and bacterial glycomes. Chem. Sci. 2010, 2, 337–344.
- García-Descalzo, L.; Alcazar, A.; Baquero, F.; Cid, C. Biotechnological Applications of Cold-Adapted Bacteria. Extremophiles: Sustainable Resources and Biotechnological Implications; Wiley-Blackwell: Hoboken, NJ, USA, 2012; pp. 159–174.
- Garcia-Lopez, E.; Rodriguez-Lorente, I.; Alcazar, P.; Cid, C. Microbial Communities in Coastal Glaciers and Tidewater Tongues of Svalbard Archipelago, Norway. Front. Mar. Sci. 2019, 5, 512.
- Garcia-Lopez, E.; Cid, C. Glaciers and Ice Sheets as Analog Environments of Potentially Habitable Icy Worlds. Front. Microbiol. 2017, 8, 1407.

