Histone deacetylase 3 is the core component of the nuclear receptor corepressor complex, and plays a central role in transcriptional repression mediated by nuclear hormone receptors.
- histone deacetylase 3
- nuclear hormone receptor
- nuclear receptor corepressor
- silencing mediator for retinoid and thyroid hormone receptors
1. Introduction
2. Basics of Molecular Action of Nuclear Hormone Receptors
NRs bind to their response elements in the DNA as heterodimers with retinoid X receptor (RXR), homodimers, or monomers, and regulate transcription of the target genes [7]. A group of NRs, including thyroid-hormone receptor (TR), retinoid A receptor (RAR), and vitamin D receptor (VDR), form heterodimers with RXR on their target genes. The N-terminal regions of these NRs are relatively small. In the absence of ligands, these NRs are associated with corepressor complexes that possess HDAC activity and suppress the transcription of the target genes, which is called ligand-independent repression [8]. Two major corepressor molecules for NRs are nuclear receptor corepressor (N-CoR) and silencing mediator for retinoid and thyroid-hormone receptors (SMRT, also known as N-CoR2). When the ligands are present, corepressors dissociate and coactivators with HAT activity are recruited instead [9] (Figure 1). This exchange is mainly due to the conformational change of NRs induced by ligands. The unliganded NRs favor binding to corepressors. Upon ligand binding, the ligand binding domain undergoes a structural transition called the “mousetrap” mechanism [10], which allows the interaction with coactivators. The cofactor exchange does not actually follow such an “all-or-none” switch model. It was revealed in vivo that the expression of TR-target genes is regulated by a shift in the relative binding of corepressors and coactivators [11]. It should be noted that these NRs similarly work as homodimers, depending on the target genes.
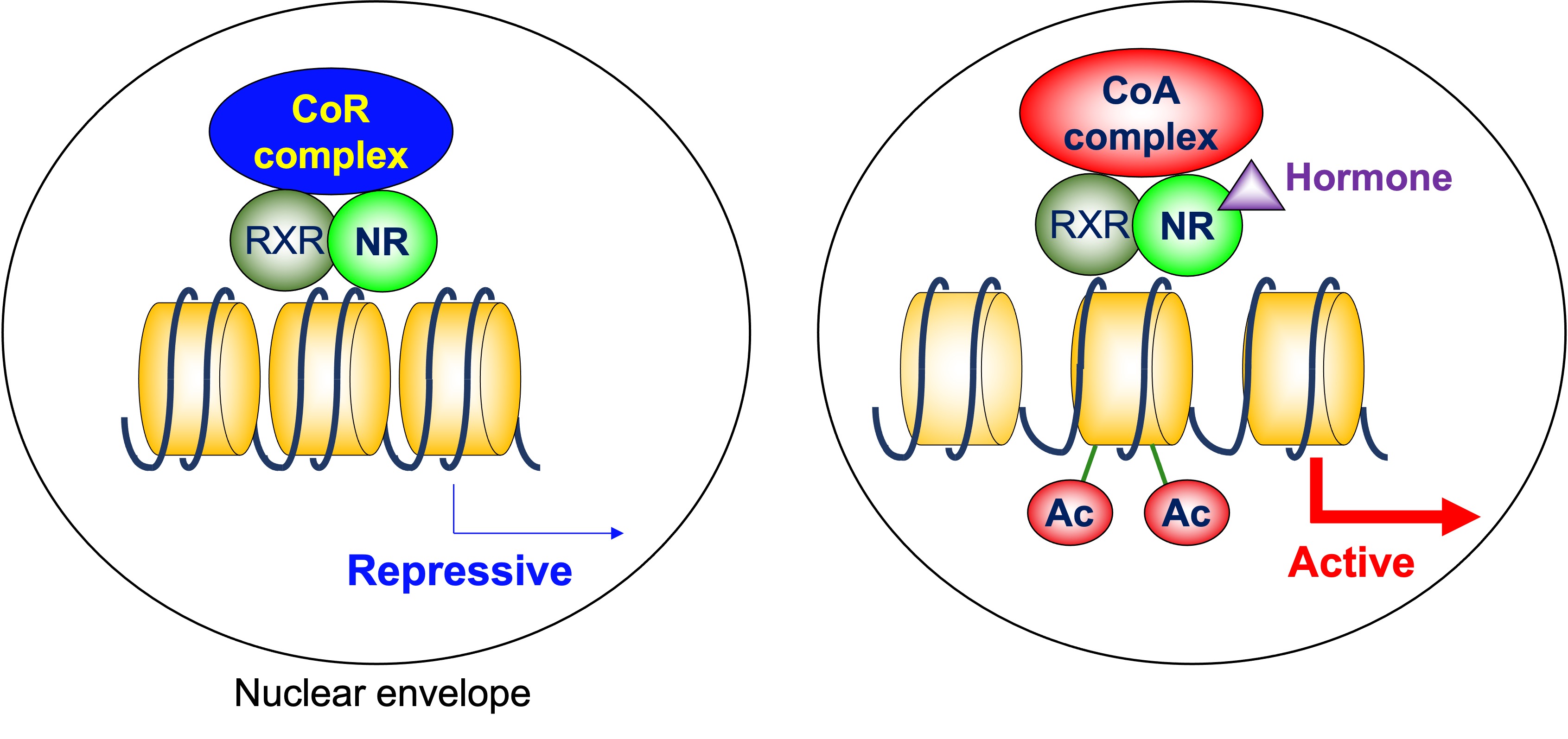
Figure 1. Cofactor exchange by heterodimeric nuclear receptors. In the absence of ligands, these NRs are associated with corepressor complexes that possess HDAC activity and suppress the transcription of the target genes, which is called ligand-independent repression. When the ligands are present, corepressors dissociate, and coactivators are recruited instead. Many of these coactivators posess HAT activity. CoR: corepressor, CoA: coactivator, NR: nuclear receptor, RXR: retinoid X receptor, Ac: acetylation.
The typical homodimerization NRs include the estrogen receptor (ER), the progesterone receptor (PR), the androgen receptor (AR), the glucocorticoid receptor (GR), and the mineralocorticoid receptor (MR). These NRs have relatively large N-terminal domains. In the absence of ligands, they bind to chaperone molecules, such as heat shock protein (HSP) 90 and HSP 70 in the cytosol [12]. In addition, unliganded ER is also localized to the nucleus, but does not bind its target genes [13]. Therefore, these NREs do not directly regulate gene transcription in the unliganded state. Upon binding to the ligands, NRs bind DNA, and stimulate transcription in association with coactivators. The conformational change by the “mousetrap” mechanism [10] is responsible for the interaction with coactivators. In this model, the role of corepressors with HDAC activity is limited.
In addition to these NRs, there are multiple orphan NRs whose ligands are yet to be identified. Whereas some of them activate transcription of the target genes in cooperation with coactivators, others serve as transcriptional repressors by interacting with corepressors.
3. Histone deacetylase 3 in nuclear hormone receptor corepressor complex
Although acetylation of histones has been extensively studied, histones are not the only substrates for acetylation and deacetylation. Given the importance of acetylation of non-histone proteins, HDACs have been renamed to lysine deacetylases (KDACs). Similarly, HATs are officially called lysine acetyltransferases (KATs). However, the familiar name “HDAC” will be used in this review.
HDAC3 was initially reported soon after HDAC1 [14][15][16]. It is ubiquitously expressed in many cell lines and tissues. Bhaskara et al. reported that HDAC3-knockout mice were embryonic lethal and HDAC3-deficient embryonic fibroblasts exhibited a delay in cell cycle progression, cell cycle-dependent DNA damage, and apoptosis [17]. Indeed, HDAC3 plays pivotal roles in maintaining basic properties of cells [18][19][20][21][22][23]. Tissue-specific functions of HDAC3 have been studied using cell-type specific knockout mice and are reviewed extensively elsewhere [24].
HDAC3 was implicated in nuclear hormone receptor action by the identification of its core interaction partners. Biochemical purification of HDAC3-associated protein complex revealed that N-CoR and SMRT are the members of the HDAC3 core complex [25][26]. Reciprocally, N-CoR and SMRT form a stable complex with HDAC3, but not with other HDACs [26][27][28][29]. These findings indicate that HDAC3 is the main HDAC responsible for the ligand-independent repression by NRs, because both N-CoR and SMRT are the major mediators of repression. These corepressors tether HDAC3 to transcriptional repressors like unliganded NRs. Moreover, interaction with N-CoR/SMRT substantially potentiates the enzymatic activity of HDAC3 [30]. Additional HDAC3 core complex members include transducin beta-like 1 X-linked (TBL1X), TBL1X receptor 1 (TBL1XR1), and G protein pathway suppressor 2 (GPS-2). These molecules also play important roles in the functions of corepressor complex [31].
4. Histone deacetylase 3 in nuclear hormone receptor action
4.1. Ligand-independent repression
As mentioned above, a heterodimeric group of NRs, including TR, RAR, and VDR, is associated with N-CoR/SMRT complexes that contain HDAC3 and suppress the transcription of the target genes in the absence of ligand. On the other hand, the unliganded homodimerization NRs, including ER, PR, GR, MR, and AR, are “neutral” in terms of gene expression because they do not bind their target genes. The findings showing the importance of ligand-independent repression include its role in phenotypic expression of TR-knockout mice and hypothyroid mice. The defects are much more severe in hypothyroid mice than in TR-deficient mice, indicating the roles of unliganded TR [32]. As a core deacetylase component of N-CoR/SMRT complex, HDAC3 plays a critical role in ligand-independent repression by deacetylating histone tails (Figure 2).
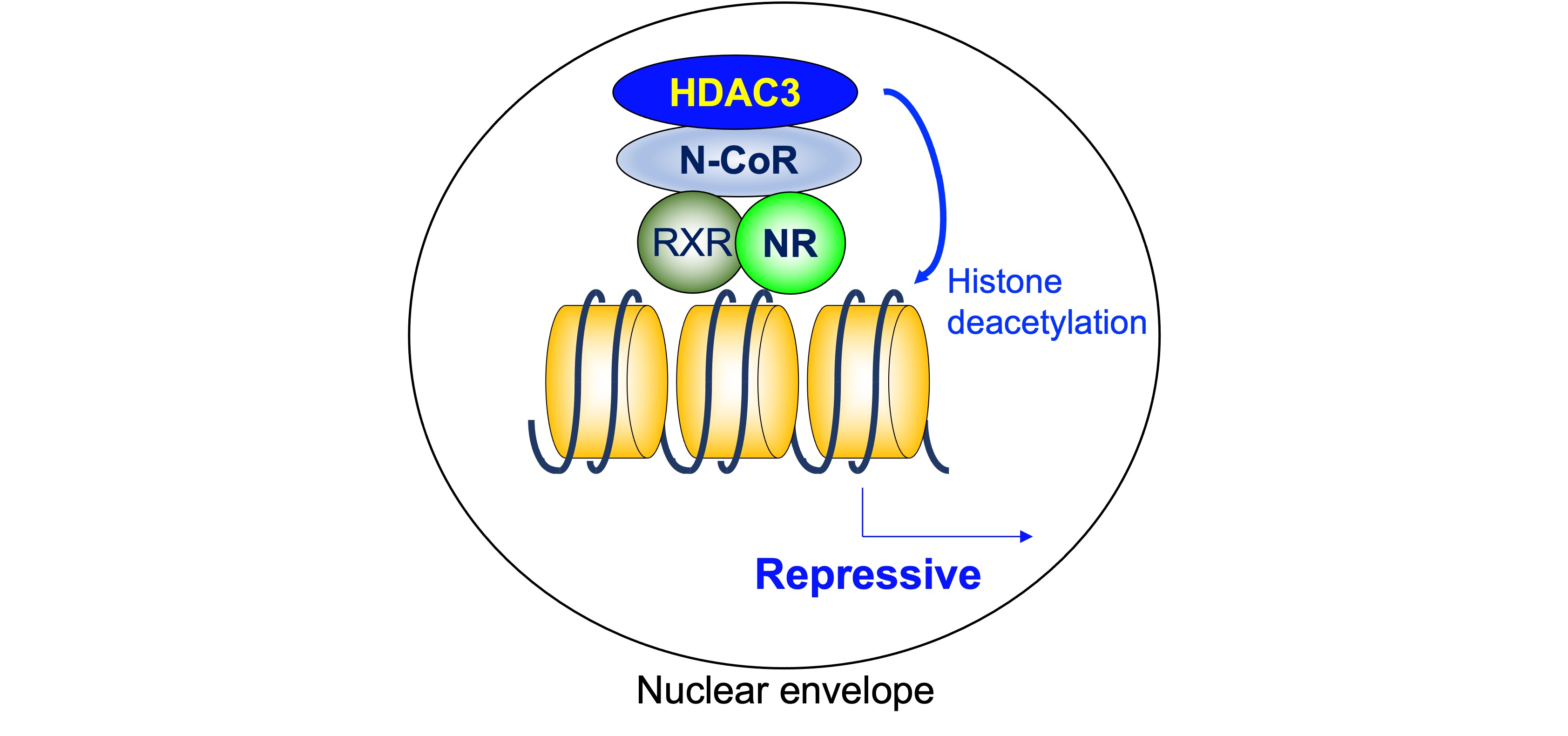
Figure 2. Histone deacetylase 3 in ligand-independent repression. As a core deacetylase component of N-CoR/SMRT complex, HDAC3 plays a critical role in ligand-independent repression by deacetylating histone tails. HDAC3: histone deacetylase 3, N-CoR: nuclear receptor corepressor, NR: nuclear receptor, RXR: retinoid X receptor.
RARα is involved in the etiology of acute promyelocytic leukemia. The majority of cases is caused by chromosomal translocations that produce promyelocytic leukemia (PML)-RAR alpha (RARA) fusion protein [33]. PML-RARA competes with RAR in a dominant-negative manner and suppresses the transcription of retinoic acid-dependent differentiation genes. The PML-RARA protein is associated with N-CoR/SMRT complex, and HDAC3 is involved in the gene repression as the main HDAC [34]. Treatment with supraphysiological doses of retinoic acid dissociates N-CoR/SMRT-HDAC3 complex from the fusion protein and induces gene expression and differentiation of the tumor cells.
4.2. Gene repression by orphan nuclear hormone receptors
Gene expression is suppressed by several orphan NR, whose ligands have not yet been identified. These NRs work as monomers, homodimers, or heterodimers with RXR or other NRs. The typical molecular mechanism is similar to ligand-independent repression, where N-CoR/SMRT-HDAC3 complex plays a pivotal role. The critical involvement of HDAC3 has been extensively studied for an orphan NR, REV-ERBα, which plays a central role in circadian rhythm by repressing core clock genes [35].
4.3. Nuclear hormone receptor antagonist action
As mentioned above, a homodimeric group of NRs does not regulate transcription in the absence of ligand, and stimulates transcription by the addition of ligands. The HDAC3 complex is not involved in that model. However, some antagonists for NRs induce gene repression. HDAC3 in the N-CoR/SMRT complex is involved in transcriptional repression induced by a modulators of ER, tamoxifen [36] (Figure 3).
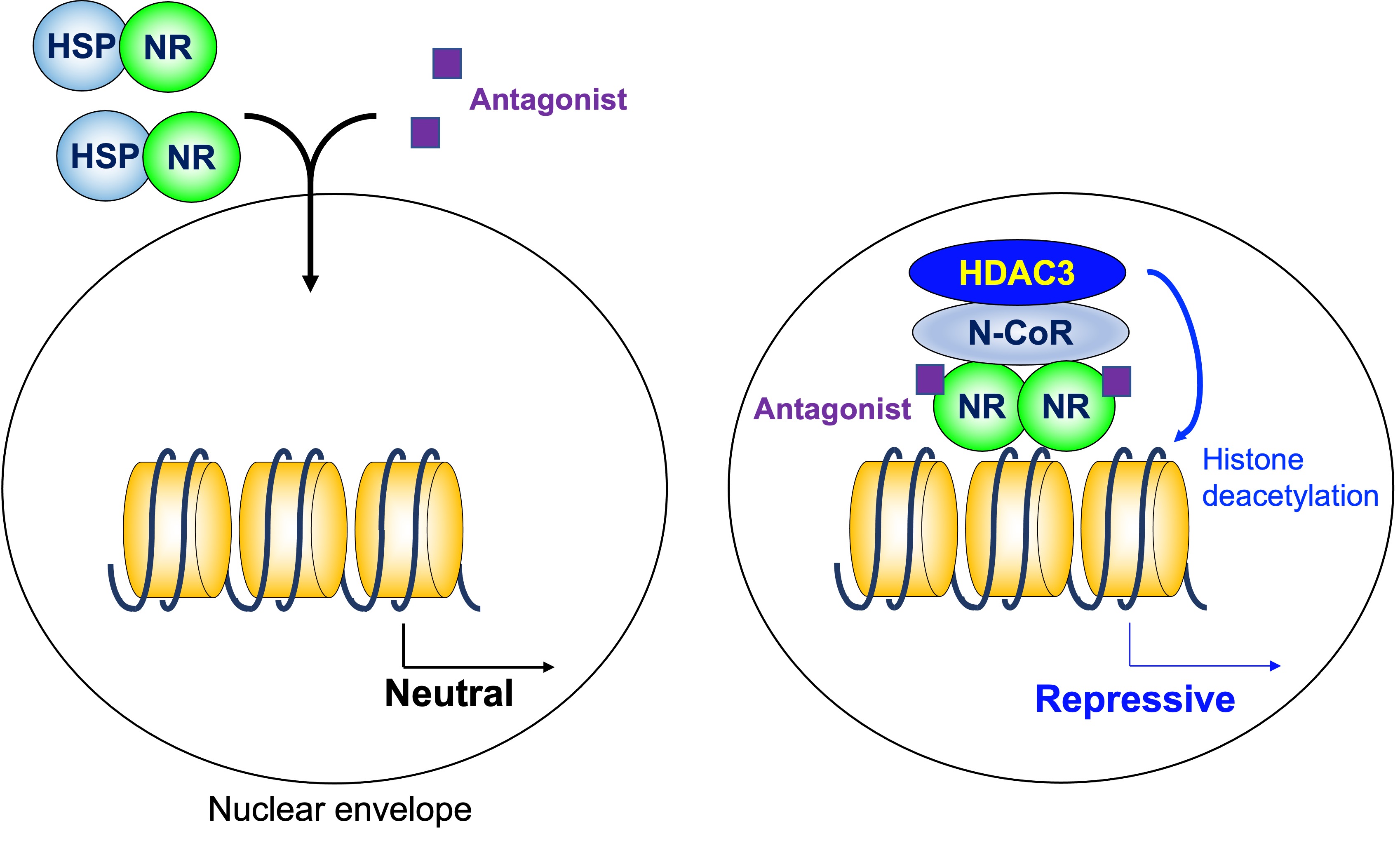
Figure 3. Histone deacetylase3 in antagonist-induced repression. Some antagonists for homodimeric NRs induce gene repression in coordination with the HDAC3 complex. HDAC3: histone deacetylase 3, N-CoR: nuclear receptor corepressor, NR: nuclear receptor, HSP: heat shock protein.
4.4. Activation of transcriptional coactivator by histone deacetylase 3
Although histone deacetylation is highly associated with gene repression, there are several genes activated by HDAC3 via NRs. At least some cases are explained by deacetylation of non-histone proteins. One convincing example is the role of HDAC3 in transcriptional stimulation in brown adipose tissue. In this model, the N-CoR/SMRT-HDAC3 complex is tethered to chromatin by the first identified orphan nuclear receptor, estrogen-related receptor α (ERRα). Peroxisome proliferator-activated receptor-γ coactivator 1α (PGC1α) is an essential coactivator for ERRα [37], and the function of PGC1α is activated by deacetylation [38]. Emmett et al. reported that HDAC3 stimulates thermogenic gene expression by deacetylating PGC1α [39].
5. Non-Canonical Mechanisms of Histone Deacetylase 3 Action for Other Transcription Factors
Although the responsible transcription factors are unknown or are not NRs, non-canonical mechanisms of HDAC3 action are reported. For example, several functions that are independent of the enzymatic activity of HDAC3 were studied. HDAC3 tethers target genes to the peripheral region of nuclei by interacting with nuclear envelope protein lamina-associated polypeptide 2 [40]. HDAC3 recruits polycomb-repressive complex 2, which possesses histone methyltransferase activity, independently of HDAC activity [41]. These tendencies suggest that non-canonical mechanisms of HDAC3 in NR action will be a hot topic in the near future. In addition, HDAC3 does not localize only in nuclei. It is also identified in the cytoplasm [42]. Therefore, it would not be surprising if cytosolic NR-HDAC3 complex exerts some transcription-independent functions.
References
- David Mangelsdorf; Carl Thummel; Miguel Beato; Peter Herrlich; Günther Schütz; Kazuhiko Umesono; Bruce Blumberg; Philippe Kastner; Manuel Mark; Pierre Chambon; et al.Ronald Evans The nuclear receptor superfamily: The second decade. Cell 1995, 83, 835-839, 10.1016/0092-8674(95)90199-x.
- David M. Lonard; Bert W. O'Malley; Nuclear receptor coregulators: modulators of pathology and therapeutic targets. Nature Reviews Endocrinology 2012, 8, 598-604, 10.1038/nrendo.2012.100.
- T. Jenuwein; Translating the Histone Code. Science 2001, 293, 1074-1080, 10.1126/science.1063127 DOI:dx.doi.org.
- Eric Verdin; Melanie Ott; 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nature Reviews Molecular Cell Biology 2014, 16, 258-264, 10.1038/nrm3931.
- David E. Sterner; Shelley L. Berger; Acetylation of Histones and Transcription-Related Factors. Microbiology and Molecular Biology Reviews 2000, 64, 435-59, 10.1128/mmbr.64.2.435-459.2000.
- Huck Hui Ng; Adrian Bird; Histone deacetylases: silencers for hire. Trends in Biochemical Sciences 2000, 25, 121-126, 10.1016/s0968-0004(00)01551-6.
- Christopher K. Glass; Differential Recognition of Target Genes by Nuclear Receptor Monomers, Dimers, and Heterodimers*. Endocrine Reviews 1994, 15, 391-407, 10.1210/edrv-15-3-391.
- Xiao Hu; Mitchell A Lazar; Transcriptional Repression by Nuclear Hormone Receptors. Trends in Endocrinology & Metabolism 2000, 11, 6-10, 10.1016/s1043-2760(99)00215-5.
- Valentina Perissi; Kristen Jepsen; Christopher K. Glass; Michael G. Rosenfeld; Deconstructing repression: evolving models of co-repressor action. Nature Reviews Genetics 2010, 11, 109-123, 10.1038/nrg2736.
- Jean-Paul Renaud; Natacha Rochel; Marc Ruff; Valéria Vivat; Pierre Chambon; Hinrich Gronemeyer; Dino Moras; Crystal structure of the RAR-γ ligand-binding domain bound to all-trans retinoic acid. Nature 1995, 378, 681-689, 10.1038/378681a0.
- Yehuda Shabtai; Nagaswaroop K. Nagaraj; Kirill Batmanov; Young-Wook Cho; Yuxia Guan; Chunjie Jiang; Jarrett Remsberg; Douglas Forrest; Mitchell A. Lazar; A coregulator shift, rather than the canonical switch, underlies thyroid hormone action in the liver. Genes & Development 2021, 35, 367-378, 10.1101/gad.345686.120.
- W. B. Pratt; Steroid Receptor Interactions with Heat Shock Protein and Immunophilin Chaperones. Endocrine Reviews 1997, 18, 306-360, 10.1210/er.18.3.306.
- Guy Leclercq; Marc Lacroix; Ioanna Laios; Guy Laurent; Estrogen Receptor Alpha: Impact of Ligands on Intracellular Shuttling and Turnover Rate in Breast Cancer Cells. Current Cancer Drug Targets 2006, 6, 39-64, 10.2174/156800906775471716.
- Wen-Ming Yang; Ya-Li Yao; Jian-Min Sun; James Davie; Edward Seto; Isolation and Characterization of cDNAs Corresponding to an Additional Member of the Human Histone Deacetylase Gene Family. Journal of Biological Chemistry 1997, 272, 28001-28007, 10.1074/jbc.272.44.28001.
- Stephane Emiliani; Wolfgang Fischle; Carine Van Lint; Yousef Al-Abed; Eric Verdin; Characterization of a human RPD3 ortholog, HDAC3. Proceedings of the National Academy of Sciences 1998, 95, 2795-2800, 10.1073/pnas.95.6.2795.
- Fernando Dangond; David A. Hafler; Jeffrey K. Tong; Jeffrey Randallc; Ryoji Kojimac; Nalân Utku; Steven R. Gullans; Differential Display Cloning of a Novel Human Histone Deacetylase (HDAC3) cDNA from PHA-Activated Immune Cells. Biochemical and Biophysical Research Communications 1998, 242, 648-652, 10.1006/bbrc.1997.8033.
- Srividya Bhaskara; Brenda J. Chyla; Joseph M. Amann; Sarah K. Knutson; David Cortez; Zu-Wen Sun; Scott W. Hiebert; Deletion of Histone Deacetylase 3 Reveals Critical Roles in S Phase Progression and DNA Damage Control. Molecular Cell 2008, 30, 61-72, 10.1016/j.molcel.2008.02.030.
- Yun Li; Gary D. Kao; Benjamin A. Garcia; Jeffrey Shabanowitz; Donald F. Hunt; Jun Qin; Caroline Phelan; Mitchell A. Lazar; A novel histone deacetylase pathway regulates mitosis by modulating Aurora B kinase activity. Genes & Development 2006, 20, 2566-2579, 10.1101/gad.1455006.
- Sumiyasu Ishii; Yasuhiro Kurasawa; Jiemin Wong; Li-Yuan Yu-Lee; Histone deacetylase 3 localizes to the mitotic spindle and is required for kinetochore-microtubule attachment. Proceedings of the National Academy of Sciences 2008, 105, 4179-4184, 10.1073/pnas.0710140105.
- Grégory Eot-Houllier; Géraldine Fulcrand; Yoshinori Watanabe; Laura Magnaghi-Jaulin; Christian Jaulin; Histone deacetylase 3 is required for centromeric H3K4 deacetylation and sister chromatid cohesion. Genes & Development 2008, 22, 2639-2644, 10.1101/gad.484108.
- Srividya Bhaskara; Sarah K. Knutson; Guochun Jiang; Mahesh Chandrasekharan; Andrew J. Wilson; Siyuan Zheng; Ashwini Yenamandra; Kimberly Locke; Jia-Ling Yuan; Alyssa R. Bonine-Summers; et al.Christina E. WellsJonathan F. KaiserM. Kay WashingtonZhongming ZhaoFlorence F. WagnerZu-Wen SunFen XiaEdward B. HolsonDineo KhabeleScott W. Hiebert Hdac3 Is Essential for the Maintenance of Chromatin Structure and Genome Stability. Cancer Cell 2010, 18, 436-447, 10.1016/j.ccr.2010.10.022.
- Alyssa R. Summers; Melissa A. Fischer; Kristy Stengel; Yue Zhao; Jonathan F. Kaiser; Christina E. Wells; Aubrey Hunt; Srividya Bhaskara; Jessica W. Luzwick; Shilpa Sampathi; et al.Xi ChenMary Ann ThompsonDavid CortezScott W. Hiebert HDAC3 is essential for DNA replication in hematopoietic progenitor cells. Journal of Clinical Investigation 2013, 123, 3112-3123, 10.1172/JCI60806.
- Yindi Jiang; Jenny Hsieh; HDAC3 controls gap 2/mitosis progression in adult neural stem/progenitor cells by regulating CDK1 levels. Proceedings of the National Academy of Sciences 2014, 111, 13541-13546, 10.1073/pnas.1411939111.
- Matthew J. Emmett; Mitchell A. Lazar; Integrative regulation of physiology by histone deacetylase 3. Nature Reviews Molecular Cell Biology 2018, 20, 102-115, 10.1038/s41580-018-0076-0.
- Yu-Der Wen; Valentina Perissi; Lena M. Staszewski; Wen-Ming Yang; Anna Krones; Christopher K. Glass; Michael G. Rosenfeld; Edward Seto; The histone deacetylase-3 complex contains nuclear receptor corepressors. Proceedings of the National Academy of Sciences 2000, 97, 7202-7207, 10.1073/pnas.97.13.7202.
- Jinsong Zhang; Markus Kalkum; Brian T Chait; Robert G Roeder; The N-CoR-HDAC3 Nuclear Receptor Corepressor Complex Inhibits the JNK Pathway through the Integral Subunit GPS2. Molecular Cell 2002, 9, 611-623, 10.1016/s1097-2765(02)00468-9.
- Matthew G. Guenther; William S. Lane; Wolfgang Fischle; Eric Verdin; Mitchell A. Lazar; Ramin Shiekhattar; A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genome Research 2000, 14, 1048-1057.
- Qiushi Lin; Specific targeting and constitutive association of histone deacetylase complexes during transcriptional repression. Genes & Development 2002, 16, 687-692, 10.1101/gad.962502.
- Ho-Geun Yoon Yoon; Doug W. Chan; Zhi‐Qing Huang; Jiwen Li; Joseph D. Fondell; Jun Qin; Jiemin Wong; Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. The EMBO Journal 2003, 22, 1336-1346, 10.1093/emboj/cdg120.
- Matthew G. Guenther; Orr Barak; Mitchell A. Lazar; The SMRT and N-CoR Corepressors Are Activating Cofactors for Histone Deacetylase 3. Molecular and Cellular Biology 2001, 21, 6091-6101, 10.1128/mcb.21.18.6091-6101.2001.
- P Karagianni; J Wong; HDAC3: taking the SMRT-N-CoRrect road to repression. Oncogene 2007, 26, 5439-5449, 10.1038/sj.onc.1210612.
- Fredric E. Wondisford; Thyroid Hormone Action: Insight from Transgenic Mouse Models. Journal of Investigative Medicine 2003, 51, 215-220, 10.1136/jim-51-04-22.
- Joaquin J. Jimenez; Ravinder S. Chale; Andrea C. Abad; Andrew V. Schally; Acute promyelocytic leukemia (APL): a review of the literature. Oncotarget 2020, 11, 992-1003, 10.18632/oncotarget.27513.
- Akihide Atsumi; Akihiro Tomita; Hitoshi Kiyoi; Tomoki Naoe; Histone deacetylase 3 (HDAC3) is recruited to target promoters by PML-RARα as a component of the N-CoR co-repressor complex to repress transcription in vivo. Biochemical and Biophysical Research Communications 2006, 345, 1471-1480, 10.1016/j.bbrc.2006.05.047.
- Yong Hoon Kim; Sajid A. Marhon; Yuxiang Zhang; David J. Steger; Kyoung-Jae Won; Mitchell A. Lazar; Rev-erbα dynamically modulates chromatin looping to control circadian gene transcription. Science 2018, 359, 1274-1277, 10.1126/science.aao6891.
- Xue-Feng Liu; Milan K. Bagchi; Recruitment of Distinct Chromatin-modifying Complexes by Tamoxifen-complexed Estrogen Receptor at Natural Target Gene Promoters in Vivo. Journal of Biological Chemistry 2004, 279, 15050-15058, 10.1074/jbc.m311932200.
- Sylvia N. Schreiber; Darko Knutti; Kathrin Brogli; Thomas Uhlmann; Anastasia Kralli; The Transcriptional Coactivator PGC-1 Regulates the Expression and Activity of the Orphan Nuclear Receptor Estrogen-Related Receptor α (ERRα). Journal of Biological Chemistry 2003, 278, 9013-9018, 10.1074/jbc.m212923200.
- Joseph Rodgers; Carlos Lerin; Wilhelm Haas; Steven P. Gygi; Bruce M. Spiegelman; Pere Puigserver; Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 2005, 434, 113-118, 10.1038/nature03354.
- Matthew Emmett; Hee-Woong Lim; Jennifer Jager; Hannah Richter; Marine Adlanmerini; Lindsey C. Peed; Erika R. Briggs; David J. Steger; Tao Ma; Carrie A. Sims; et al.Joseph A. BaurLiming PeiKyoung Jae WonPatrick SealeZachary Gerhart-HinesMitchell A. Lazar Histone deacetylase 3 prepares brown adipose tissue for acute thermogenic challenge. Nature 2017, 546, 544-548, 10.1038/nature22819.
- Raz Somech; Sigal Shaklai; Orit Geller; Ninette Amariglio; Amos J. Simon; Gideon Rechavi; Einav Nili Gal-Yam; The nuclear-envelope protein and transcriptional repressor LAP2β interacts with HDAC3 at the nuclear periphery, and induces histone H4 deacetylation. Journal of Cell Science 2005, 118, 4017-4025, 10.1242/jcs.02521.
- Sara Lewandowski; Harish Palleti Janardhan; Chinmay M. Trivedi; Histone Deacetylase 3 Coordinates Deacetylase-independent Epigenetic Silencing of Transforming Growth Factor-β1 (TGF-β1) to Orchestrate Second Heart Field Development. Journal of Biological Chemistry 2015, 290, 27067-27089, 10.1074/jbc.m115.684753.
- Yasunari Takami; Tatsuo Nakayama; N-terminal Region, C-terminal Region, Nuclear Export Signal, and Deacetylation Activity of Histone Deacetylase-3 Are Essential for the Viability of the DT40 Chicken B Cell Line. Journal of Biological Chemistry 2000, 275, 16191-16201, 10.1074/jbc.m908066199.

