Mitochondria are double membrane-bound organelles consisting of an outer membrane, intermembrane space (IMS), inner membrane, and matrix that play critical functions in cells including metabolism, energy production, regulation of intrinsic apoptosis, and maintenance of calcium homeostasis. Mitochondria are fascinatingly equipped with their own genome and machinery for transcribing and translating 13 essential proteins of the oxidative phosphorylation system (OXPHOS). The rest of the proteins (99%) that function in mitochondria in the various pathways described above are nuclear-transcribed and synthesized as precursors in the cytosol. These proteins are imported into the mitochondria by the unique mitochondrial protein import system that consists of seven machineries. Proper functioning of the mitochondrial protein import system is crucial for optimal mitochondrial deliverables, as well as mitochondrial and cellular homeostasis.
- mitochondria
- outer membrane
- proteins
1. Introduction
Mitochondria are organelles present in almost all eukaryotic cells, and their number per cell depends on their energy demand. Organs with high metabolic activity, for example, heart muscles, kidneys, and the brain, contain the largest number of mitochondria [1,2]. Mitochondria are believed to be the descendants of an ancient prokaryote that underwent an endosymbiotic event with early eukaryotes [3]. Apart from their role in energy production, mitochondria are involved in numerous metabolic processes, including the biosynthesis of amino acids, lipids, heme, and Fe-S clusters [4]. In addition, they also play crucial functions in programmed cell death and maintenance of calcium homeostasis [4,5]. A mitochondrion is a double membrane-bound structure consisting of an outer membrane, intermembrane space (IMS), inner membrane, and matrix [6]. The outer membrane is permeable to solutes up to approximately 5 kDa and characterized by the presence of various enzymes and channels, such as carnitine palmitoyltransferase I, acyl-CoA synthetase, voltage-dependent anion channel (VDAC), and mitochondrial apoptosis-induced channel (MAC) [7]. On the other hand, the inner membrane is impermeable except through specific transporters and contains the enzyme complexes responsible for oxidative phosphorylation [6,8]. The IMS consists of enzymes including caspases, adenylyl kinase, and cytochrome c. Similarly, the mitochondrial matrix consists of several enzymes that take part in metabolic processes such as the tricarboxylic acid cycle and β oxidation of fatty acids.
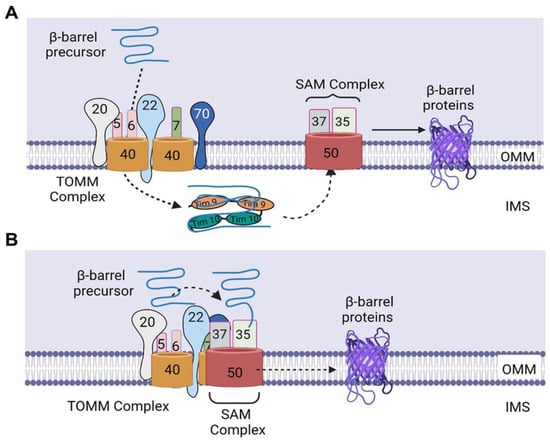
Each mitochondrion has its own genome, which is a 16.5 kb double-stranded, closed circular DNA present in the mitochondrial matrix. The mtDNA is strictly maternally inherited and packaged into nucleoids, which is done to ensure its proper distribution and propagation [9]. The mitochondrial genome consists of 37 genes that encode approximately 1% of the total mitochondrial proteins (13 OXPHOS proteins), 2 ribosomal RNAs (12S and 16S rRNA), and 22 transfer RNAs [6]. The remaining 99% of mitochondrial proteins (~1500) are encoded by the nuclear genome, synthesized in the cytosol, and imported into the mitochondria by the mitochondrial protein import system, which consists of seven machineries (Figure 1). The translocase of the outer mitochondrial membrane (TOMM) machinery appears to be the most important of these machineries, as it is the first to come in contact with majority of the nuclear-encoded mitochondrial proteins to allow their entry into the intermembrane space. Subsequently, these proteins make use of any of the other machineries to get to their final destination. The choice of the next machinery depends on the targeting signal and the destination of the protein.

Figure 1. The mitochondrial protein import system. The mitochondrial protein import system consists of seven machineries, including the translocase of the outer mitochondrial membrane (TOMM) machinery, mitochondrial import machinery (MIM), sorting and assembly machinery (SAM), mitochondrial intermembrane space import and assembly machinery (MIA), translocase of the inner mitochondrial membrane 23 (TIMM 23) machinery, translocase of the inner mitochondrial membrane 22 (TIMM 22) machinery, and a presequence-associated motor (PAM). The figure was created with Biorender.com.
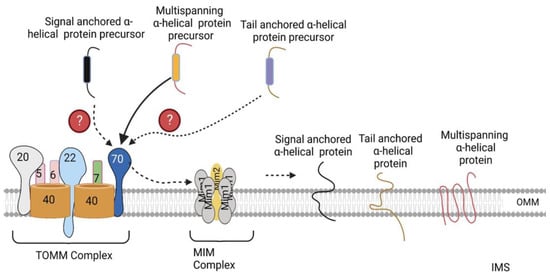
The mitochondrial outer membrane possesses two types of integral membrane proteins, including β-barrel proteins that are integrated into the OMM by multiple transmembrane β strands, and α-helical proteins, which are anchored in the OMM by one or more hydrophobic α-helical segments [4,10].
2. Sorting of β-Barrel Proteins into the Outer Mitochondrial Membrane
3. Sorting of α-Helical Proteins into the Outer Mitochondrial Membrane
This entry is adapted from the peer-reviewed paper 10.3390/ijms24032479
References
- Wang, Z.; Ying, Z.; Bosy-Westphal, A.; Zhang, J.; Schautz, B.; Later, W.; Heymsfield, S.B.; Müller, M.J. Specific metabolic rates of major organs and tissues across adulthood: Evaluation by mechanistic model of resting energy expenditure. Am. J. Clin. Nutr. 2010, 92, 1369–1377.
- Pagliarini, D.J.; Calvo, S.E.; Chang, B.; Sheth, S.A.; Vafai, S.B.; Ong, S.-E.; Walford, G.A.; Sugiana, C.; Boneh, A.; Chen, W.K.; et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell 2008, 134, 112–123.
- Archibald, J.M. Endosymbiosis and Eukaryotic Cell Evolution. Curr. Biol. 2015, 25, R911–R921.
- Wiedemann, N.; Pfanner, N. Mitochondrial Machineries for Protein Import and Assembly. Annu Rev Biochem 2017, 86, 685–714.
- SSrinivasan; Guha, M.; Kashina, A.; Avadhani, N.G. Mitochondrial dysfunction and mitochondrial dynamics-The cancer connection. Biochim. Biophys. Acta Bioenerg. 2017, 1858, 602–614.
- Adebayo, M.; Singh, S.; Singh, A.P.; Dasgupta, S. Mitochondrial fusion and fission: The fine-tune balance for cellular homeostasis. FASEB J. 2021, 35, e21620.
- O’Rourke, B. Mitochondrial ion channels. Annu. Rev. Physiol. 2007, 69, 19–49.
- Lemasters, J.J. Modulation of mitochondrial membrane permeability in pathogenesis, autophagy and control of metabolism. J. Gastroenterol. Hepatol. 2007, 22, S31–S37.
- Gilkerson, R.W. Mitochondrial DNA nucleoids determine mitochondrial genetics and dysfunction. Int. J. Biochem. Cell Biol. 2009, 41, 1899–1906.
- Chacinska, A.; Koehler, C.M.; Milenkovic, D.; Lithgow, T.; Pfanner, N. Importing mitochondrial proteins: Machineries and mechanisms. Cell 2009, 138, 628–644.
- Dudek, J.; Rehling, P.; van der Laan, M. Mitochondrial protein import: Common principles and physiological networks. Biochim. Biophys. Acta 2013, 1833, 274–285.
- Wang, W.; Chen, X.; Zhang, L.; Yi, J.; Ma, Q.; Yin, J.; Zhuo, W.; Gu, J.; Yang, M. Atomic structure of human TOM core complex. Cell Discov. 2020, 6, 67.
- Jores, T.; Klinger, A.; Groß, L.E.; Kawano, S.; Flinner, N.; Duchardt-Ferner, E.; Wöhnert, J.; Kalbacher, H.; Endo, T.; Schleiff, E.; et al. Characterization of the targeting signal in mitochondrial β-barrel proteins. Nat. Commun. 2016, 7, 12036.
- Qiu, J.; Wenz, L.-S.; Zerbes, R.M.; Oeljeklaus, S.; Bohnert, M.; Stroud, D.A.; Wirth, C.; Ellenrieder, L.; Thornton, N.; Kutik, S.; et al. Coupling of mitochondrial import and export translocases by receptor-mediated supercomplex formation. Cell 2013, 154, 596–608.
- Krimmer, T.; Rapaport, D.; Ryan, M.; Meisinger, C.; Kassenbrock, C.K.; Blachly-Dyson, E.; Forte, M.; Douglas, M.G.; Neupert, W.; Nargang, F.E.; et al. Biogenesis of porin of the outer mitochondrial membrane involves an import pathway via receptors and the general import pore of the TOM complex. J. Cell Biol. 2001, 152, 289–300.
- Wiedemann, N.; Truscott, K.N.; Pfannschmidt, S.; Guiard, B.; Meisinger, C.; Pfanner, N. Biogenesis of the protein import channel Tom40 of the mitochondrial outer membrane: Intermembrane space components are involved in an early stage of the assembly pathway. J. Biol. Chem. 2004, 279, 18188–18194.
- Kutik, S.; Stojanovski, D.; Becker, L.; Becker, T.; Meinecke, M.; Krüger, V.; Prinz, C.; Meisinger, C.; Guiard, B.; Wagner, R.; et al. Dissecting membrane insertion of mitochondrial beta-barrel proteins. Cell 2008, 132, 1011–1024.
- Diederichs, K.A.; Ni, X.; Rollauer, S.E.; Botos, I.; Tan, X.; King, M.S.; Kunji, E.R.S.; Jiang, J.; Buchanan, S.K. Structural insight into mitochondrial β-barrel outer membrane protein biogenesis. Nat. Commun. 2020, 11, 3290.
- Wenz, L.S.; Ellenrieder, L.; Qiu, J.; Bohnert, M.; Zufall, N.; van der Laan, M.; Pfanner, N.; Wiedemann, N.; Becker, T. Sam37 is crucial for formation of the mitochondrial TOM-SAM supercomplex, thereby promoting β-barrel biogenesis. J. Cell Biol. 2015, 210, 1047–1054.
- Ahting, U.; Waizenegger, T.; Neupert, W.; Rapaport, D. Signal-anchored proteins follow a unique insertion pathway into the outer membrane of mitochondria. J. Biol. Chem. 2005, 280, 48–53.
- Meineke, B.; Engl, G.; Kemper, C.; Vasiljev-Neumeyer, A.; Paulitschke, H.; Rapaport, D. The outer membrane form of the mitochondrial protein Mcr1 follows a TOM-independent membrane insertion pathway. FEBS Lett. 2008, 582, 855–860.
- Becker, T.; Wenz, L.-S.; Krüger, V.; Lehmann, W.; Müller, J.M.; Goroncy, L.; Zufall, N.; Lithgow, T.; Guiard, B.; Chacinska, A.; et al. The mitochondrial import protein Mim1 promotes biogenesis of multispanning outer membrane proteins. J. Cell Biol. 2011, 194, 387–395.
- Papic, D.; Krumpe, K.; Dukanovic, J.; Dimmer, K.S.; Rapaport, D. Multispan mitochondrial outer membrane protein Ugo1 follows a unique Mim1-dependent import pathway. J. Cell Biol. 2011, 194, 397–405.
- Doan, K.N.; Grevel, A.; Mårtensson, C.U.; Ellenrieder, L.; Thornton, N.; Wenz, L.-S.; Opaliński, .; Guiard, B.; Pfanner, N.; Becker, T. The Mitochondrial Import Complex MIM Functions as Main Translocase for α-Helical Outer Membrane Proteins. Cell Rep. 2020, 31, 107567.
- Kemper, C.; Habib, S.; Engl, G.; Heckmeyer, P.; Dimmer, K.S.; Rapaport, D. Integration of tail-anchored proteins into the mitochondrial outer membrane does not require any known import components. J. Cell Sci. 2008, 121, 1990–1998.
- Vögtle, F.-N.; Keller, M.; Taskin, A.A.; Horvath, S.E.; Guan, X.L.; Prinz, C.; Opalińska, M.; Zorzin, C.; van der Laan, M.; Wenk, M.R.; et al. The fusogenic lipid phosphatidic acid promotes the biogenesis of mitochondrial outer membrane protein Ugo1. J. Cell Biol. 2015, 210, 951–960.
- Sinzel, M.; Tan, T.; Wendling, P.; Kalbacher, H.; Özbalci, C.; Chelius, X.; Westermann, B.; Brügger, B.; Rapaport, D.; Dimmer, K.S. Mcp3 is a novel mitochondrial outer membrane protein that follows a unique IMP-dependent biogenesis pathway. EMBO Rep. 2016, 17, 965–981.
