Corn silk (CS) is an agro-by-product from corn cultivation. It is used in folk medicines in some countries, besides being commercialized as health-promoting supplements and beverages. Unlike CS-derived natural products, their bioactive peptides, particularly antioxidant peptides, are understudied.
- antioxidant
- in silico
- in vitro
- mechanism
- molecular docking
- peptide
- purification
- Stigma maydis
1. Introduction
Image by M Ameen from Pixabay
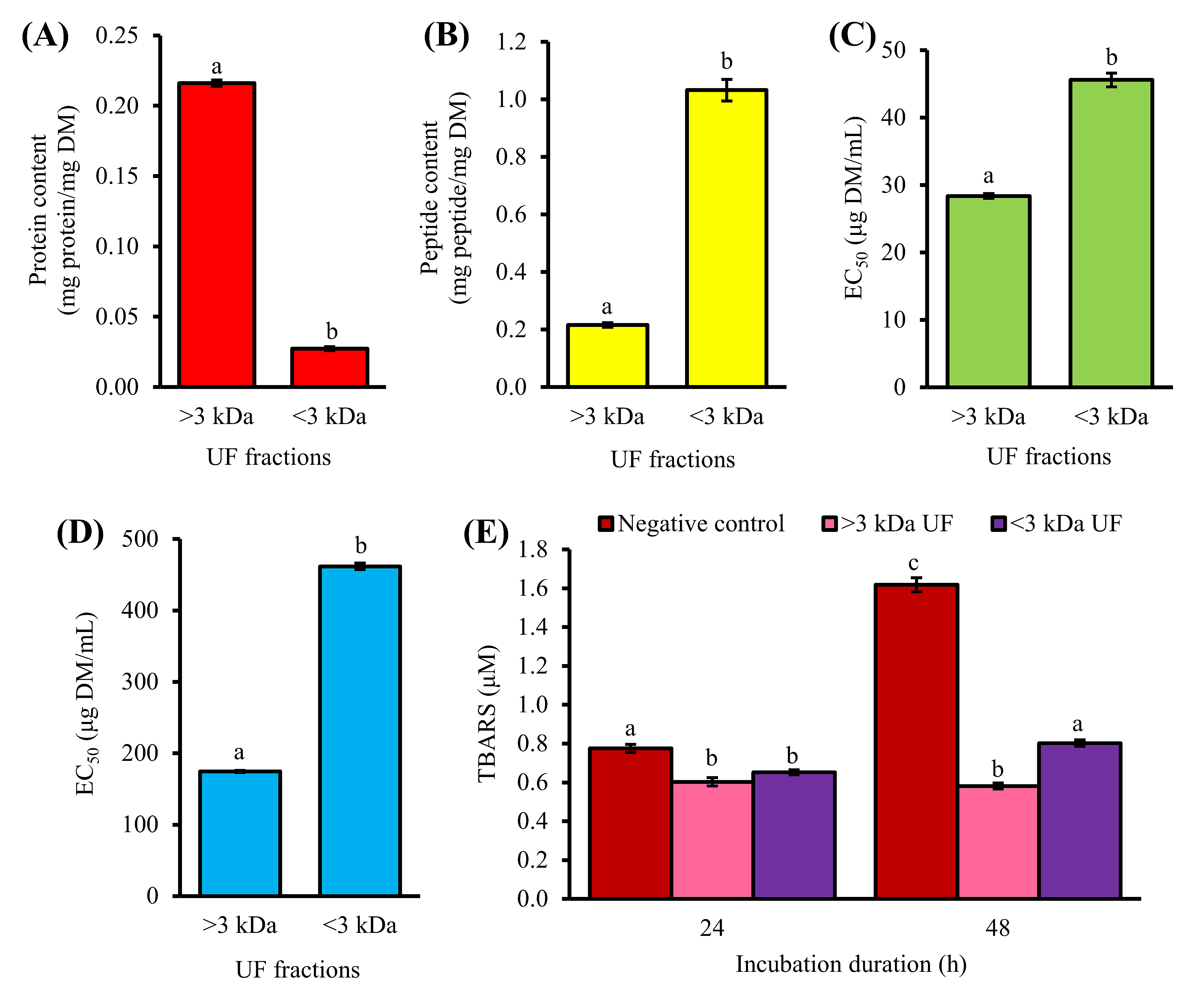
2. Purification of 1-h Trypsin Hydrolysate by Ultrafiltration
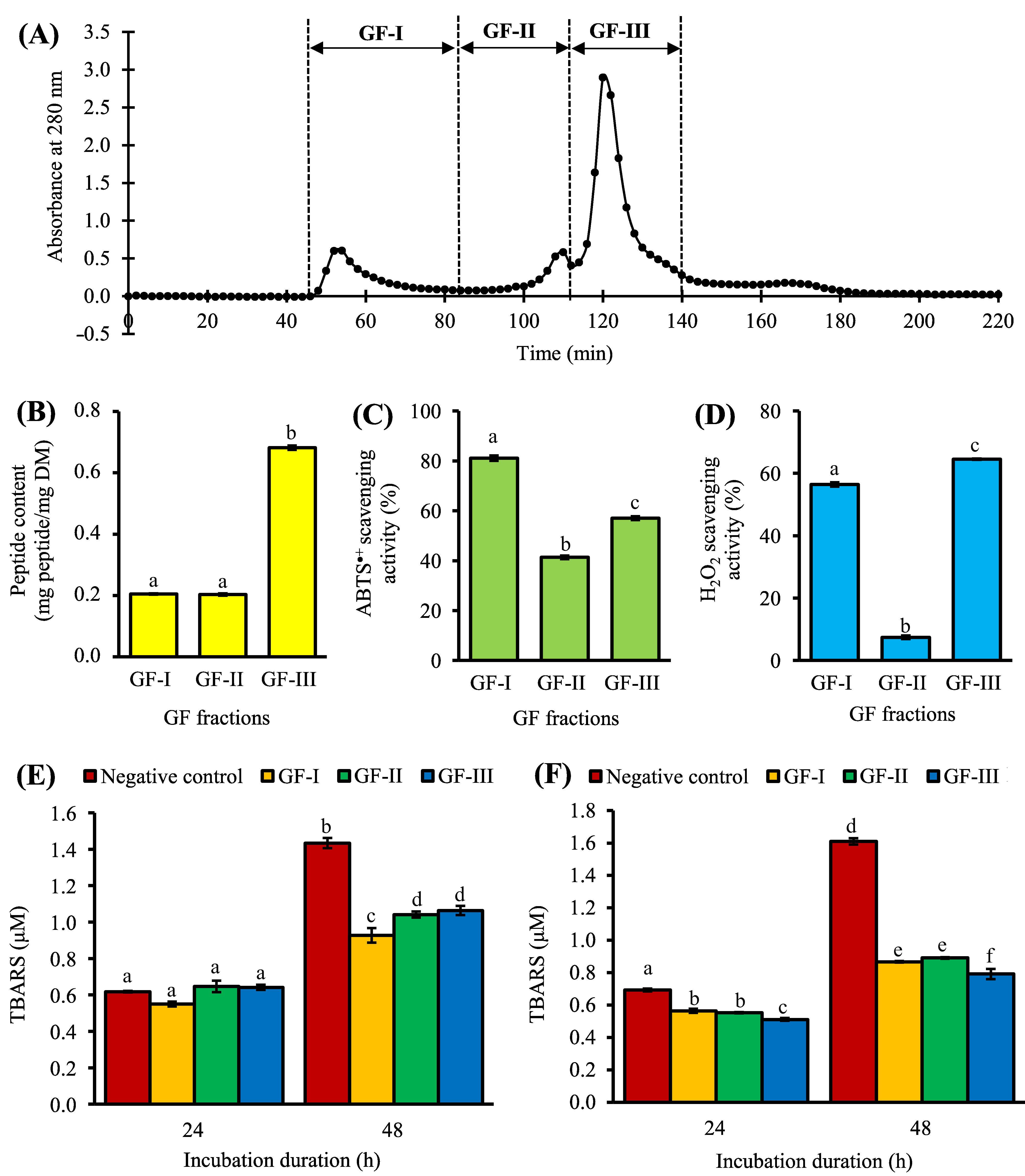
3. Purification of <3 kDa Fraction by Gel Filtration Chromatography
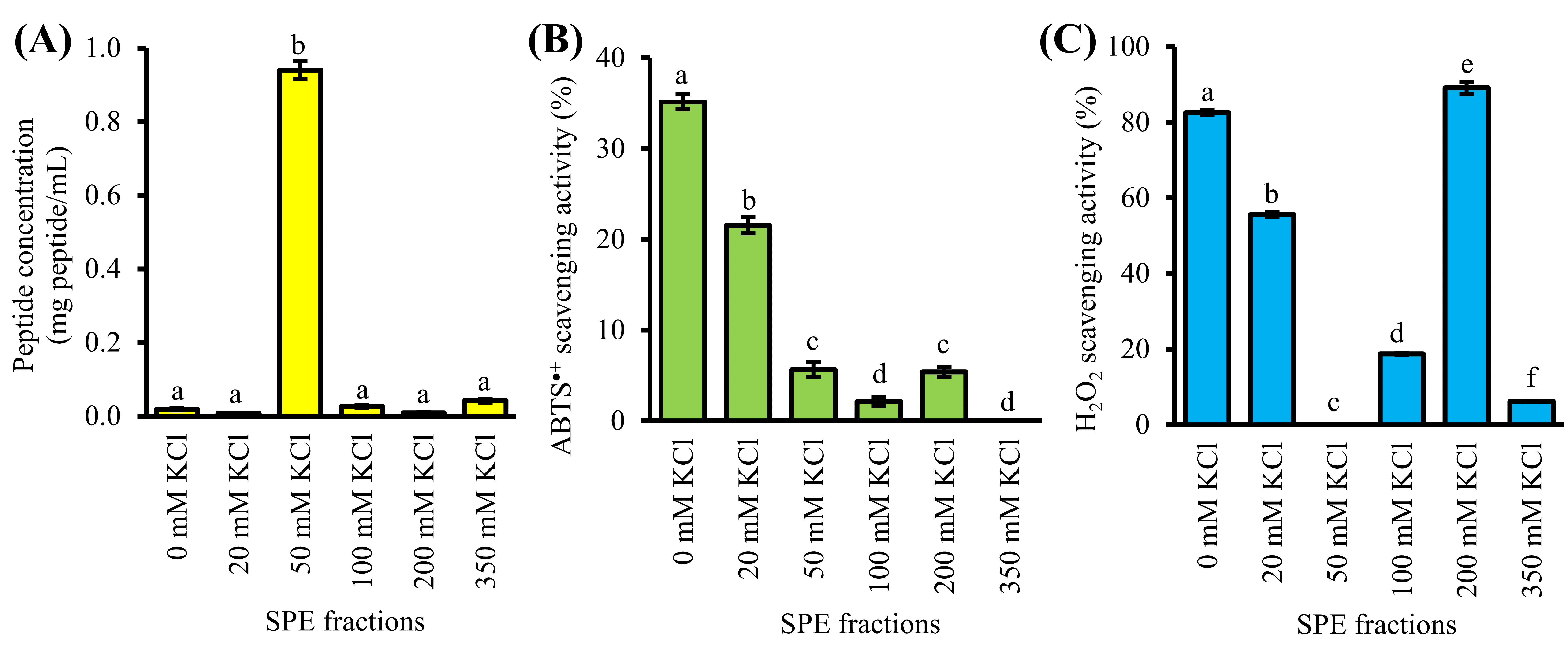
4. Purification of GF-III by Strong-Cation-Exchange Solid-Phase Extraction
5. Identification and Characterization of Antioxidant Peptides
| SPE Fractions | Peptides | Measured m/z [M + 2H]2 |
Molecular Mass (Da) a |
Aromatic Residues (%) b |
Basic Residues (%) b |
Hydrophobic Residues (%) b |
Aliphatic Residues (%) b |
Aliphatic Index b |
|---|---|---|---|---|---|---|---|---|
| 0 mM KCl | KRYFKR | 449.28 | 896.57 | 33 | 67 | 33 | 0 | 0 |
| PRVRVAGR | 455.79 | 909.58 | 0 | 38 | 63 | 38 | 85 | |
| PVVWAAKR | 463.79 | 925.57 | 13 | 25 | 75 | 50 | 98 | |
| QVASGPLQR | 478.28 | 954.55 | 0 | 11 | 56 | 33 | 87 | |
| MAPRTPRK | 478.78 | 955.57 | 0 | 38 | 50 | 13 | 13 | |
| NKVVKLMR | 494.31 | 986.62 | 0 | 38 | 50 | 38 | 121 | |
| KVPLAVFSR | 508.82 | 1015.64 | 11 | 22 | 67 | 44 | 119 | |
| LKKGSPLKR | 513.84 | 1025.69 | 0 | 44 | 44 | 22 | 87 | |
| FQLKPVFR | 517.82 | 1033.63 | 25 | 25 | 63 | 25 | 85 | |
| THAVKGVVHK | 538.34 | 1074.67 | 20 | 40 | 50 | 40 | 97 | |
| YTWKFKGR | 543.31 | 1084.61 | 38 | 38 | 50 | 0 | 0 | |
| ARVPQQSYR | 552.80 | 1103.61 | 11 | 22 | 44 | 22 | 43 | |
| VHFNKGKKR | 557.34 | 1112.69 | 22 | 56 | 33 | 11 | 32 | |
| TAPLSSKALKR | 586.37 | 1170.73 | 0 | 27 | 45 | 36 | 89 | |
| FSCPLVMKGPNGLR | 759.91 | 1517.81 | 7 | 14 | 71 | 21 | 76 | |
| 20 mM KCl | RHGSGR | 335.18 | 668.37 | 17 | 50 | 33 | 0 | 0 |
| NMVPGR | 337.17 | 672.34 | 0 | 17 | 67 | 17 | 48 | |
| FMFFVYK | 491.25 | 980.50 | 57 | 14 | 86 | 14 | 41 | |
| MCFHHHFHK | 612.27 | 1222.53 | 67 | 56 | 44 | 0 | 0 | |
| 200 mM KCl | DFPGAK | 317.66 | 633.33 | 17 | 17 | 67 | 17 | 17 |
| NDGPSR | 323.15 | 644.29 | 0 | 17 | 33 | 0 | 0 | |
| AGFPLGK | 345.20 | 688.41 | 14 | 14 | 86 | 29 | 70 | |
| AMQQDK | 360.66 | 719.32 | 0 | 17 | 33 | 17 | 17 | |
| NLEGYR | 376.19 | 750.38 | 17 | 17 | 50 | 17 | 65 | |
| YETLNR | 398.20 | 794.41 | 17 | 17 | 33 | 17 | 65 | |
| MPPKSTR | 408.72 | 815.43 | 0 | 29 | 43 | 0 | 0 | |
| TAGASLVAR | 423.25 | 844.49 | 0 | 11 | 67 | 56 | 109 | |
| SSPATGGSLR | 466.74 | 931.49 | 0 | 10 | 50 | 20 | 49 | |
| NANSLAGPQR | 514.27 | 1026.55 | 0 | 10 | 50 | 30 | 59 |
| Peptides | SPE Fractions | FRS Scores |
|---|---|---|
| MCFHHHFHK | 20 mM KCl | 0.68068 |
| VGPWQK * | - | 0.52254 |
| MYPGLA * | - | 0.49386 |
| NLEGYR | 200 mM KCl | 0.48158 |
| AGFPLGK | 200 mM KCl | 0.44866 |
| FMFFVYK | 20 mM KCl | 0.44397 |
| NMVPGR | 20 mM KCl | 0.44319 |
| PVVWAAKR | 0 mM KCl | 0.43744 |
| DFPGAK | 200 mM KCl | 0.43574 |
| FPLPSF * | - | 0.43352 |
| FSCPLVMKGPNGLR | 0 mM KCl | 0.41864 |
| WAFAPA * | - | 0.41519 |
| RHGSGR | 20 mM KCl | 0.41088 |
| VHFNKGKKR | 0 mM KCl | 0.41055 |
| NANSLAGPQR | 200 mM KCl | 0.40415 |
| QVASGPLQR | 0 mM KCl | 0.40213 |
| MAPRTPRK | 0 mM KCl | 0.39973 |
| NDGPSR | 200 mM KCl | 0.38760 |
| KRYFKR | 0 mM KCl | 0.38352 |
| YETLNR | 200 mM KCl | 0.37938 |
| FQLKPVFR | 0 mM KCl | 0.37599 |
| ARVPQQSYR | 0 mM KCl | 0.37580 |
| YTWKFKGR | 0 mM KCl | 0.36769 |
| AMQQDK | 200 mM KCl | 0.36324 |
| SSPATGGSLR | 200 mM KCl | 0.35382 |
| THAVKGVVHK | 0 mM KCl | 0.35200 |
| MPPKSTR | 200 mM KCl | 0.33529 |
| LKKGSPLKR | 0 mM KCl | 0.32957 |
| PRVRVAGR | 0 mM KCl | 0.32698 |
| KVPLAVFSR | 0 mM KCl | 0.32525 |
| TAGASLVAR | 200 mM KCl | 0.32285 |
| TAPLSSKALKR | 0 mM KCl | 0.29320 |
| NKVVKLMR | 0 mM KCl | 0.27437 |
6. Molecular Docking between CS Peptides and ABTS•+
| Peptides | SPE Fractions | Binding Affinity (kcal/mol) |
Peptide Residues Interacting with ABTS•+ a | |
|---|---|---|---|---|
| Hydrogen Bond | Hydrophobic Interaction | |||
| MCFHHHFHK | 20 mM KCl | −4.8 | - | Phe3, His6, Phe7 |
| VHFNKGKKR | 0 mM KCl | −4.7 | Lys7, Arg9 | Val1, His2, Gly6, Lys7, Arg9 |
| PVVWAAKR | 0 mM KCl | −4.7 | Arg8 (2) | Val2, Trp4, Ala5, Ala6, Arg8 |
| FMFFVYK | 20 mM KCl | −4.4 | Lys7 | Phe1, Phe3, Phe4, Lys7 |
| FSCPLVMKGPNGLR | 0 mM KCl | −4.2 | Arg14 (2) | Leu5, Lys8, Gly9, Pro10, Gly12, Arg14 |
| NMVPGR | 20 mM KCl | −4.1 | Asn1, Arg6 (2) | Asn1, Pro4, Gly5, Arg6 |
| NLEGYR | 200 mM KCl | −4.1 | - | Tyr5, Arg6 |
| RHGSGR | 20 mM KCl | −3.9 | Arg1, Arg6 | Arg1, Gly5, Arg6 |
| AGFPLGK | 200 mM KCl | −3.7 | - | Phe3, Pro4, Leu5 |
| DFPGAK | 200 mM KCl | −3.6 | - | Pro3, Gly4, Lys6 |
| FPLPSF * | - | −4.6 | Phe1, Ser5 | Phe1, Pro2, Leu3, Pro4, Ser5 |
| WAFAPA * | - | −4.3 | - | Trp1, Ala4, Pro5 |
| VGPWQK * | - | −3.9 | - | Pro3, Trp4, Lys6 |
| MYPGLA * | - | −3.8 | Pro3 | Pro3, Leu5, Ala6 |
| Peptides a | Basic Residues | Mutant Peptides | Binding Affinity (kcal/mol) |
|---|---|---|---|
| MCFHHHFHK | His6 | MCFHHAFHK | −4.1 |
| VHFNKGKKR | His2 | VAFNKGKKR | −4.8 |
| Lys7 | VHFNKGAKR | −5.0 | |
| Arg9 | VHFNKGKKA | −4.4 | |
| PVVWAAKR | Arg8 | PVVWAAKA | −4.3 |
| FMFFVYK | Lys7 | FMFFVYA | −4.3 |
| FSCPLVMKGPNGLR | Lys8 | FSCPLVMAGPNGLR | −4.2 |
| Arg14 | FSCPLVMKGPNGLA | −4.7 | |
| NMVPGR | Arg6 | NMVPGA | −3.1 |
| NLEGYR | Arg6 | NLEGYA | −3.7 |
| RHGSGR | Arg1 | AHGSGR | −3.8 |
| Arg6 | RHGSGA | −4.2 | |
| DFPGAK | Lys6 | DFPGAA | −3.4 |
7. Molecular Docking of Peptides on Keap1
| Peptides | Toxicity | Allergenicity | CPP Prediction |
|---|---|---|---|
| NDGPSR | Non-toxin | Probable non-allergen | CPP |
| NLEGYR | Non-toxin | Probable non-allergen | CPP |
| NMVPGR | Non-toxin | Probable non-allergen | CPP |
| SSPATGGSLR | Non-toxin | Probable non-allergen | CPP |
| NANSLAGPQR | Non-toxin | Probable non-allergen | CPP |
| KRYFKR | Non-toxin | Probable non-allergen | CPP |
| RHGSGR | Non-toxin | Probable non-allergen | CPP |
| YETLNR | Non-toxin | Probable non-allergen | Non-CPP |
| AGFPLGK | Non-toxin | Probable non-allergen | Non-CPP |
| KVPLAVFSR | Non-toxin | Probable non-allergen | Non-CPP |
| TAGASLVAR | Non-toxin | Probable allergen | Non-CPP |
| YTWKFKGR | Non-toxin | Probable allergen | CPP |
| AMQQDK | Non-toxin | Probable allergen | CPP |
| MPPKSTR | Non-toxin | Probable allergen | CPP |
| PVVWAAKR | Non-toxin | Probable allergen | CPP |
| DFPGAK | Non-toxin | Probable allergen | Non-CPP |
| FMFFVYK | Non-toxin | Probable allergen | Non-CPP |
| QVASGPLQR | Non-toxin | Probable allergen | Non-CPP |
| DEQIPSHPPR * | Non-toxin | Probable allergen | Non-CPP |
| DTETGVPT * | Non-toxin | Probable non-allergen | Non-CPP |
| VPY * | Non-toxin | Probable allergen | CPP |
| ACECD * | Non-toxin | Probable allergen | CPP |
| Peptides | Binding Affinity (kcal/mol) |
Interaction with Keap1 a | ||
|---|---|---|---|---|
| Hydrogen Bond | Hydrophobic Interaction | Salt Bridge | ||
| NLEGYR | −8.7 | Arg415, Arg483, Ser508, Gln530, Ser555 | Tyr334, Ser363, Gly364, Leu365, Ala366, Arg415, Ile416, Gly417, Gly462, Phe478, Arg483, Ser508, Gly509, Ala510, Tyr525, Gln530, Ser555, Ala556, Leu557, Tyr572, Phe577, Ser602, Gly603, Val604 | Arg415 |
| NANSLAGPQR | −8.2 | Arg415 (3), Val418, Val465, Arg483 | Ser363, Gly364, Leu365, Arg380, Asn382, Asn414, Arg415, Ile416, Gly417, Ile461, Gly462, Val463, Val465, Phe478, Arg483, Ser508, Gly509, Tyr525, Gln530, Ser555, Ala556, Ile559, Phe577, Gly603 | - |
| NMVPGR | −8.1 | Ser363, Leu365, Asn382, Ser602 | Tyr334, Ser363, Gly364, Leu365, Ala366, Asn382, Arg415, Ile416, Ile461, Gly462, Ser508, Gly509, Ala510, Tyr525, Gln530, Ser555, Ala556, Ser602 | - |
| SSPATGGSLR | −8.1 | Ser363, Arg380, Asn414, Arg415, Ser431, Ser602 | Tyr334, Gly364, Leu365, Arg380, Asn382, Asn414, Arg415, Ile416, Ser431, Gly433, His436, Gly462, Phe478, Arg483, Ser508, Gly509, Ala556, Ser602, Gly603 | - |
| NDGPSR | −8.0 | Arg415 (2), Ala510 | Tyr334, Gly364, Leu365, Arg415, Ile461, Gly462, Phe478, Ser508, Gly509, Tyr525, Ala556, Ser602, Gly603, Val604 | - |
| DEQIPSHPPR * | −8.0 | Tyr334, Asn414, Arg415 (4), Ser431, Arg483 (3), Ser555 | Tyr334, Ser363, Arg380, Asn382, Asn414, Arg415, Ser431, Gly433, His436, Gly462, Phe478, Arg483, Ser508, Gly509, Tyr525, Ser555, Ala556, Tyr572, Phe577, Ser602 | Arg483 (2) |
8. Molecular Docking of Peptides on MPO
In this study, CS peptides were compared with the soy tripeptide VPY and false abalone-derived DTETGVPT in their affinity towards MPO. Oral administration of VPY was shown to induce a 4-fold reduction in the MPO activity of mice [22]. Meanwhile, DTETGVPT is a potential inhibitor of MPO, as revealed by molecular docking simulation [23]. Our results revealed that 12 of the 29 CS peptides had up to 23% stronger affinity to MPO when compared with DTETGVPT, but were all weaker than VPY (Table S2). Five of the 12 potential MPO-binding CS peptides, namely NDGPSR, NLEGYR, NMVPGR, KRYFKR, and RHGSGR, were found to have low toxicity, low allergenicity, and cell-penetrating potential (Table 5). The five CS peptides would thus have an advantage over the reference peptide VPY (probable allergen) and DTETGVPT (non-cell-penetrating peptide) (Table 5) in the context of the application as food additives or functional food ingredients.
The interactions between the five CS peptides and MPO active site residues were highly similar to those between the reference peptides (VPY and DTETGVPT) and MPO (Table 7). With the exemption of NLEGYR, CS peptides and reference peptides were all forming only the hydrophobic interaction with the key residues in MPO active site. Likewise, similar to the reference peptides, each CS peptide could interact with 4–5 of the seven key residues in the active site of MPO. These key residues contributed to the stability of the interaction between 7-benzyl-1H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, the bound inhibitor in the MPO crystal, and the active site of MPO [24]. As observed in the two reference peptides, all the five CS peptides could bind directly to the heme moiety of MPO (Table 7). Blockage of the heme pocket of MPO could preclude the access of substrates to the MPO active site, suppressing the enzyme’s action-driving ROS production [25].
Table 7. Binding affinities and types of interactions between myeloperoxidase (MPO) and five corn silk peptides predicted as non-toxic, non-allergenic and cell-penetrating peptides, in comparison with two reference peptides.
| Peptides | Binding Affinity (kcal/mol) |
Interaction with MPO a | ||
|---|---|---|---|---|
| Hydrogen Bond | Hydrophobic Interaction | Salt Bridge | ||
| NMVPGR | -6.6 | - | Phe99, Thr100, Glu102, Glu116, Pro145, Phe147, Leu216, Pro220, Arg239, Glu242, Phe366, Phe407, Met411, Arg424, Hec606 | - |
| NLEGYR | -6.5 | His95 | His95, Phe99, Glu102, Glu116, Pro145, Phe146, Phe147, Pro220, Thr238, Arg239, Glu242, Phe407, Val410, Met411, Leu420, Hec606 | - |
| NDGPSR | -6.3 | Glu102 | Phe99, Glu102, Glu116, Pro145, Phe146, Phe147, Pro220, Thr238, Arg239, Glu242, Phe366, Phe407, Met411, Leu415, Leu420, Hec606 | - |
| RHGSGR | -6.2 | Thr100, Thr238 | Phe99, Thr100, Glu102, Pro145, Phe146, Phe147, Leu216, Pro220, Thr238, Arg239, Glu242, Phe366, Phe407, Met411, Leu415, Hec606 | Glu102 (5) |
| KRYFKR | -5.5 | Thr100, Thr238 | His95, Phe99, Thr100, Glu102, Glu116, Pro145, Phe147, Pro220, Thr238, Arg239, Glu242, Phe366, Phe407, Val410, Met411, Leu415, Leu420, Hec606 | Glu102 (2) |
| VPY * | -7.4 | - | His95, Phe99, Thr100, Glu102, Pro220, Thr238, Arg239, Glu242, Phe366, Hec606 | - |
| DTETGVPT * | -5.5 | Thr238 | Phe99, Thr100, Glu102, Phe147, Pro220, Thr238, Arg239, Glu242, Phe366, Phe407, Met411, Leu415, Leu420, Hec606 | - |
* Indicates reference peptides. a Number in brackets indicates the number of interactions. Residues in bold are key residues (Gln91, His95, Phe99, Arg239, His336, Phe366, and Phe407) in the active site of MPO [24].
9. Molecular Docking of Peptides on XO
Only 14 of the 29 CS peptides were docked onto XO successfully, yielding negative values for binding affinity (Table S2). Docking of NDGPSR and DFPGAK onto XO involved the lowest binding energy, thus the strongest affinity, among the 14 peptides. Furthermore, NDGPSR and DFPGAK were predicted to bind equally stably to XO as was ACECD, the reference peptide. ACECD, an XO inhibitory peptide derived from Skipjack tuna hydrolysate, was reported to exert its inhibition by binding to the active site of XO [26]. In light of the predicted allergenicity and non-cell-penetrating potential of DFPGAK (Table 5), we proceeded to analyze the intermolecular interactions in only the NDGPSR-XO docked model.
As shown in Table 8, our LigPlot+ analysis revealed that NDGPSR could interact with catalytically critical residues (Glu802 and Arg880), substrate binding-residue (Phe914, Phe1009, and Thr1010), and the residues associated with the extended solvent-accessible channel leading to the molybdenum active center (Leu873, Val1011, Phe1013, and Leu1014) [27]. Most of such interactions are hydrophobic in nature. The dominance of hydrophobic interactions was also observed in our ACECD-XO docked model (Table 8). Meanwhile, NDGPSR could form hydrophobic interaction with Glu1261, an amino acid around the active center of XO, and contribute to the catalytic reaction of XO [26]. Moreover, as observed in ACECD, NDGPSR could also interact with Leu648 and Phe649 through hydrophobic interactions. The two residues are at the gate of the aforementioned solvent-accessible channel [27]. Interactions with Leu648 and Phe649 were reported to enhance the potency of quercetin as an XO inhibitor [27]. Altogether, our results suggest that NDGPSR can potentially occupy the catalytic center of XO, hindering the entry of XO substrates, thereby suppressing XO activity and ROS generation. In addition to their protective role in cells, antioxidant peptides with XO inhibitory activity are useful in reducing milk-fat oxidation in the dairy industry [28]. Thus, NDGPSR may also be developed into a potent antioxidant for the formulation of dairy products and other oxidation-prone foods.
Table 8. Binding affinity and types of interactions between xanthine oxidase (XO) and corn silk peptide NDGPSR, which was predicted to be a non-toxic, non-allergenic, and cell-penetrating peptide, in comparison with a reference peptide.
| Peptides | Binding Affinity (kcal/mol) |
Interaction with XO a | ||
|---|---|---|---|---|
| Hydrogen Bond | Hydrophobic Interaction | Salt Bridge | ||
| NDGPSR | -5.2 | Ser876, Thr1010, Val1011 | Leu648, Phe649, Gly799, Glu802, Leu873, His875, Ser876, Arg880, Phe914, Phe1009, Thr1010, Val1011, Pro1012, Phe1013, Leu1014, Ala1078, Ala1079, Glu1261 | His875, Glu1261 (2) |
| ACECD * | -5.2 | His875, Ser876 | Leu648, Phe649, Glu802, Leu873, His875, Ser876, Glu879, Phe914, Phe1009, Thr1010, Val1011, Pro1012, Phe1013, Leu1014 | - |
* Indicates reference peptide. a Number in brackets indicates the number of interactions. Residues in bold are key residues (Glu802, Leu873, Arg880, Phe914, Phe1009, Thr1010, Val1011, Phe1013, and Leu1014) in the active site of XO [27].
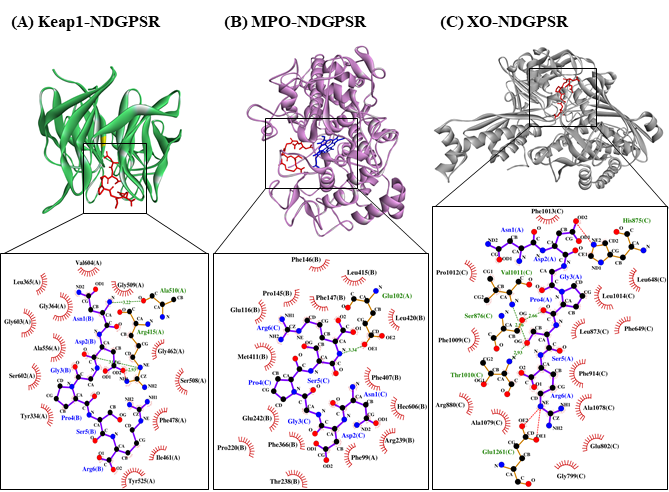
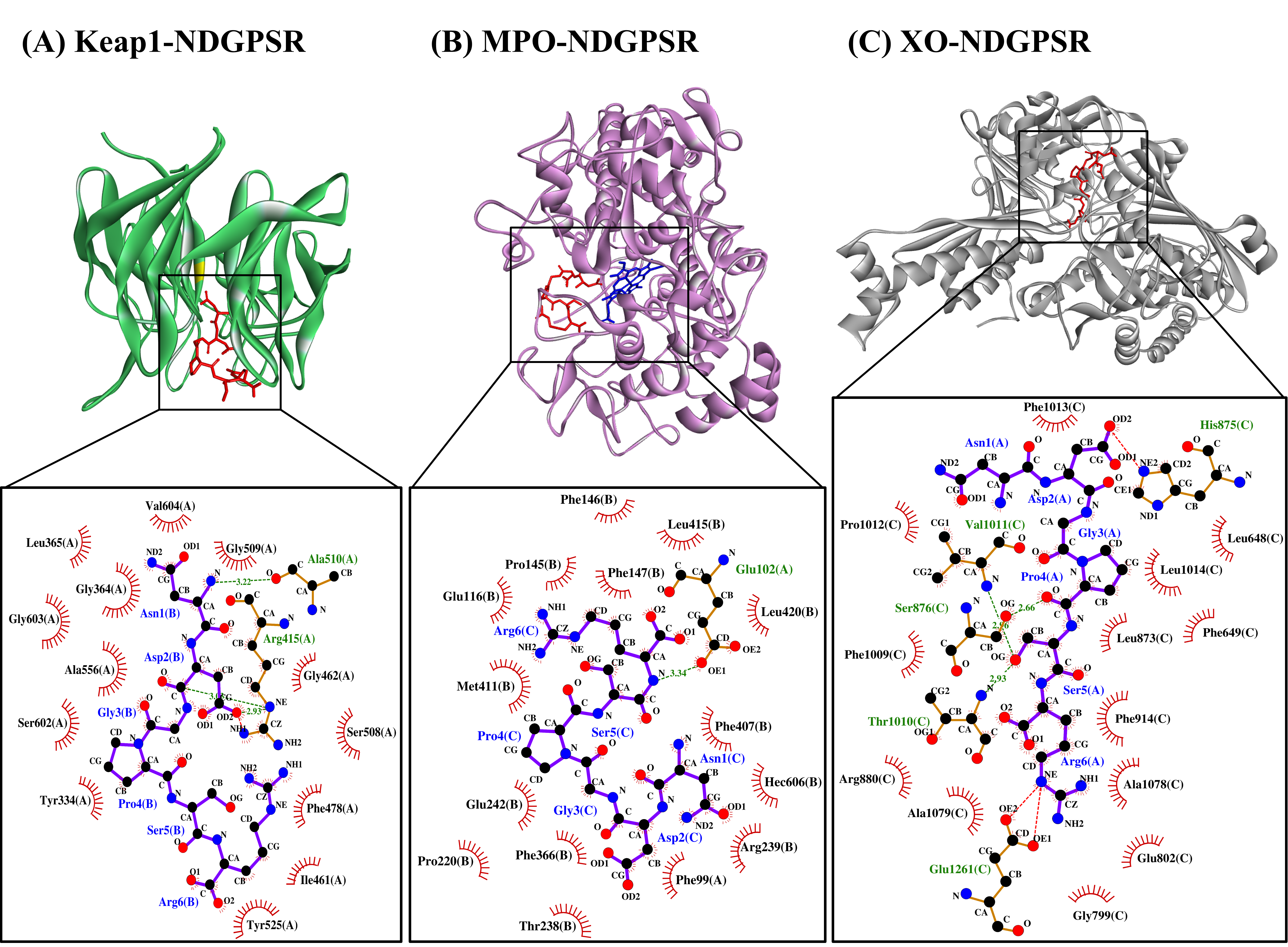
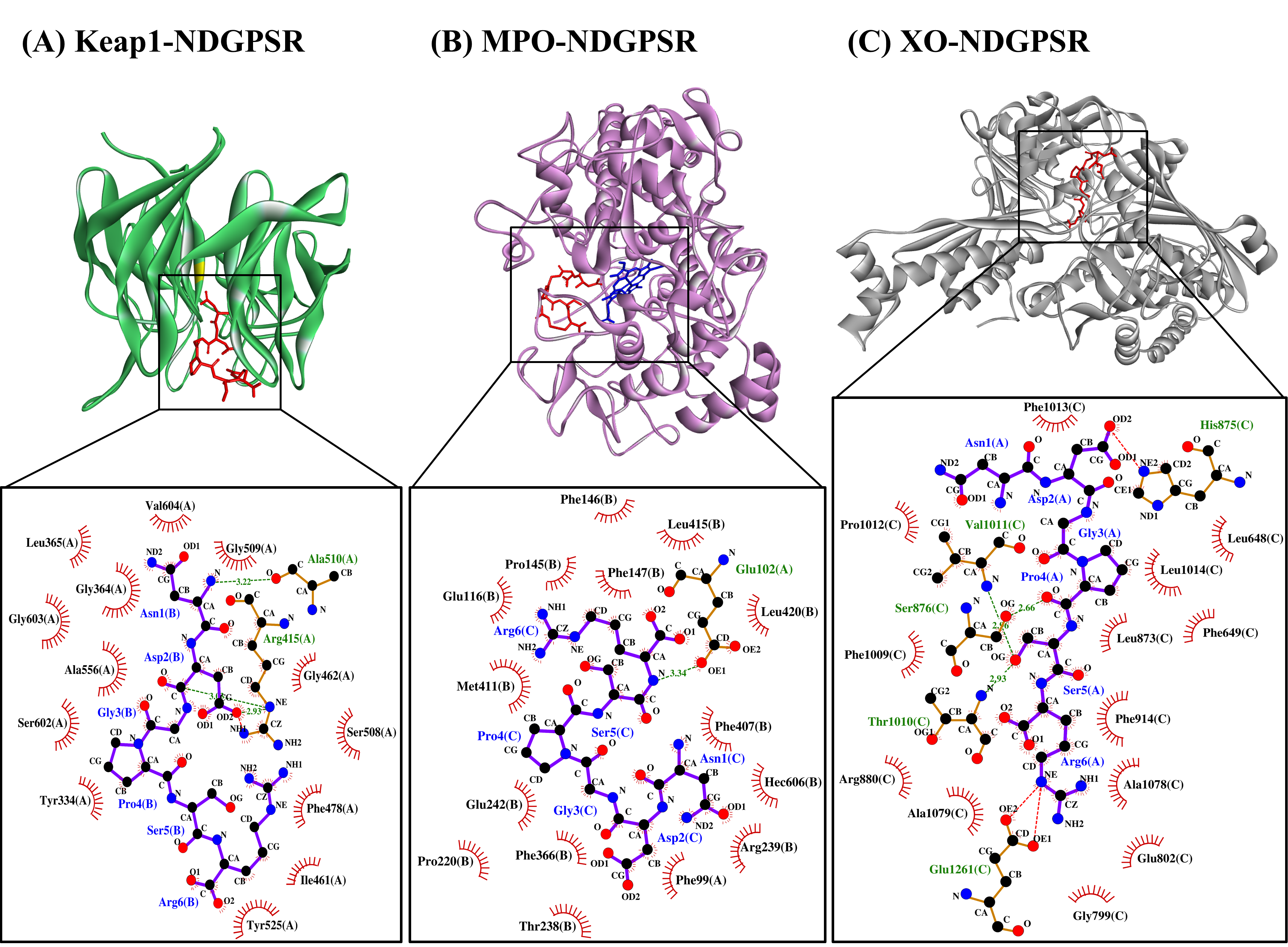
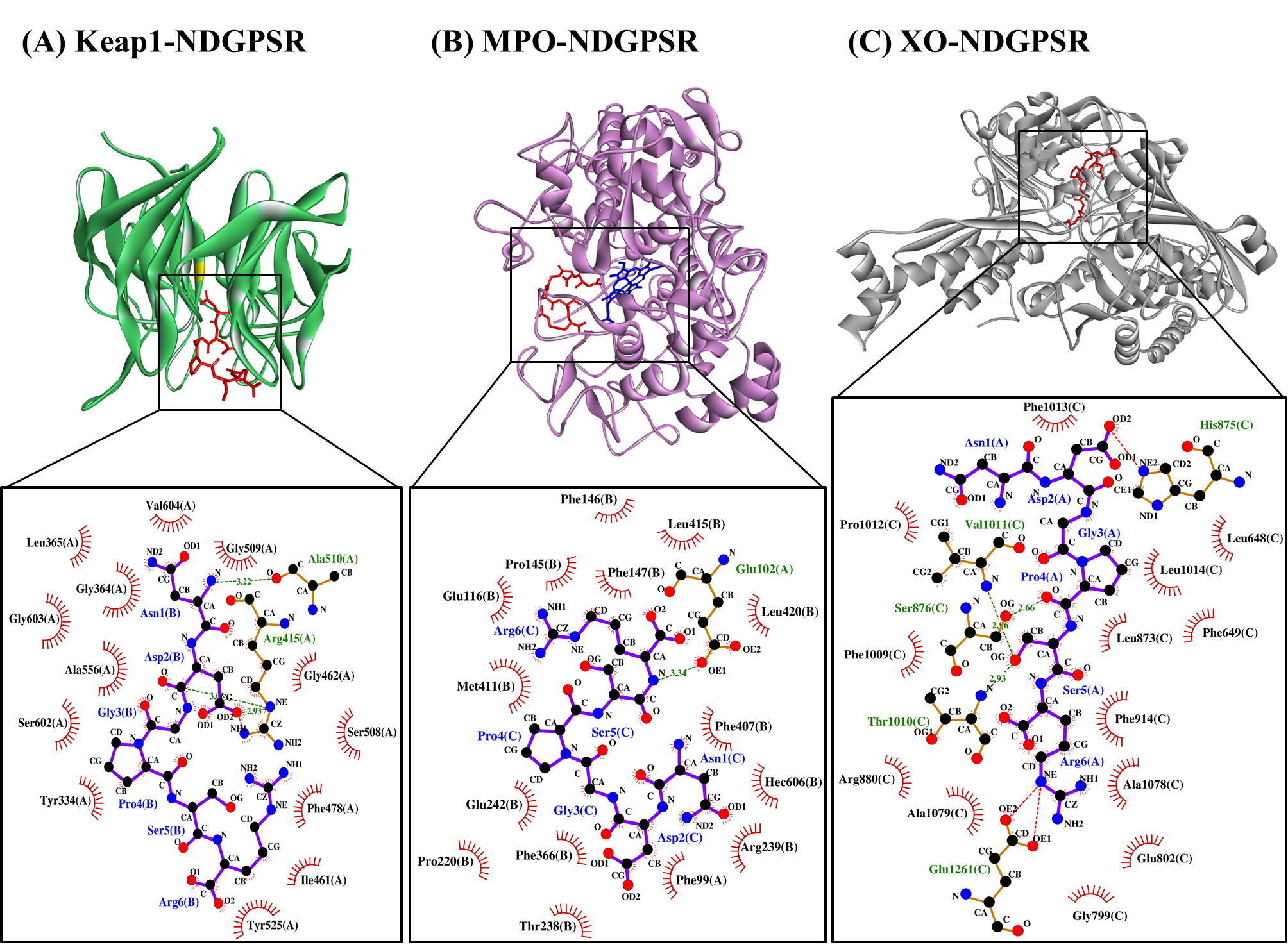
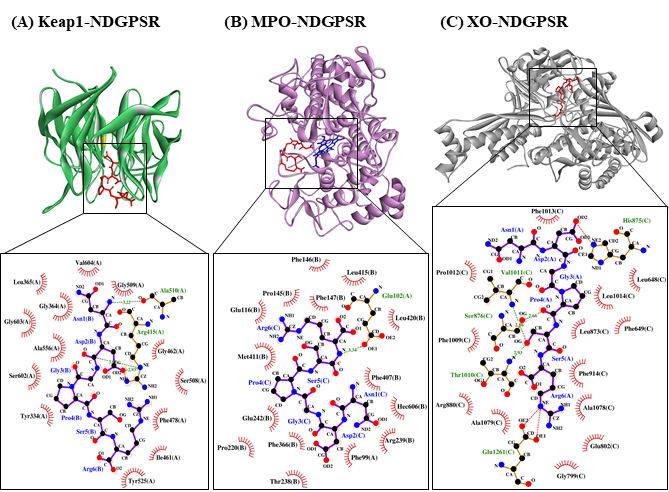
Our in silico analysis revealed three multifunctional peptides that are also non-toxic, non-allergenic, and have cell-penetrating potential, namely NDGPSR, NLEGYR, and NMVPGR. Among the three, NDGPSR could be a potential inhibitor of Keap1-Nrf2 interaction, MPO, and XO. Models of NDGPSR docked to the three protein targets are shown in Figure 4. Moreover, NLEGYR and NMVPGR could be potential dual-function inhibitors of Keap1-Nrf2 interaction and MPO (Table S2). Antioxidant peptides possessing multiple functionalities likely have greater versatility and commercial value when compared to other antioxidant peptides [29]. In this context, the three multifunctional peptides, with their safety and cell-penetrating properties, are desirable candidates for the future development of functional food ingredients and/or health supplements.
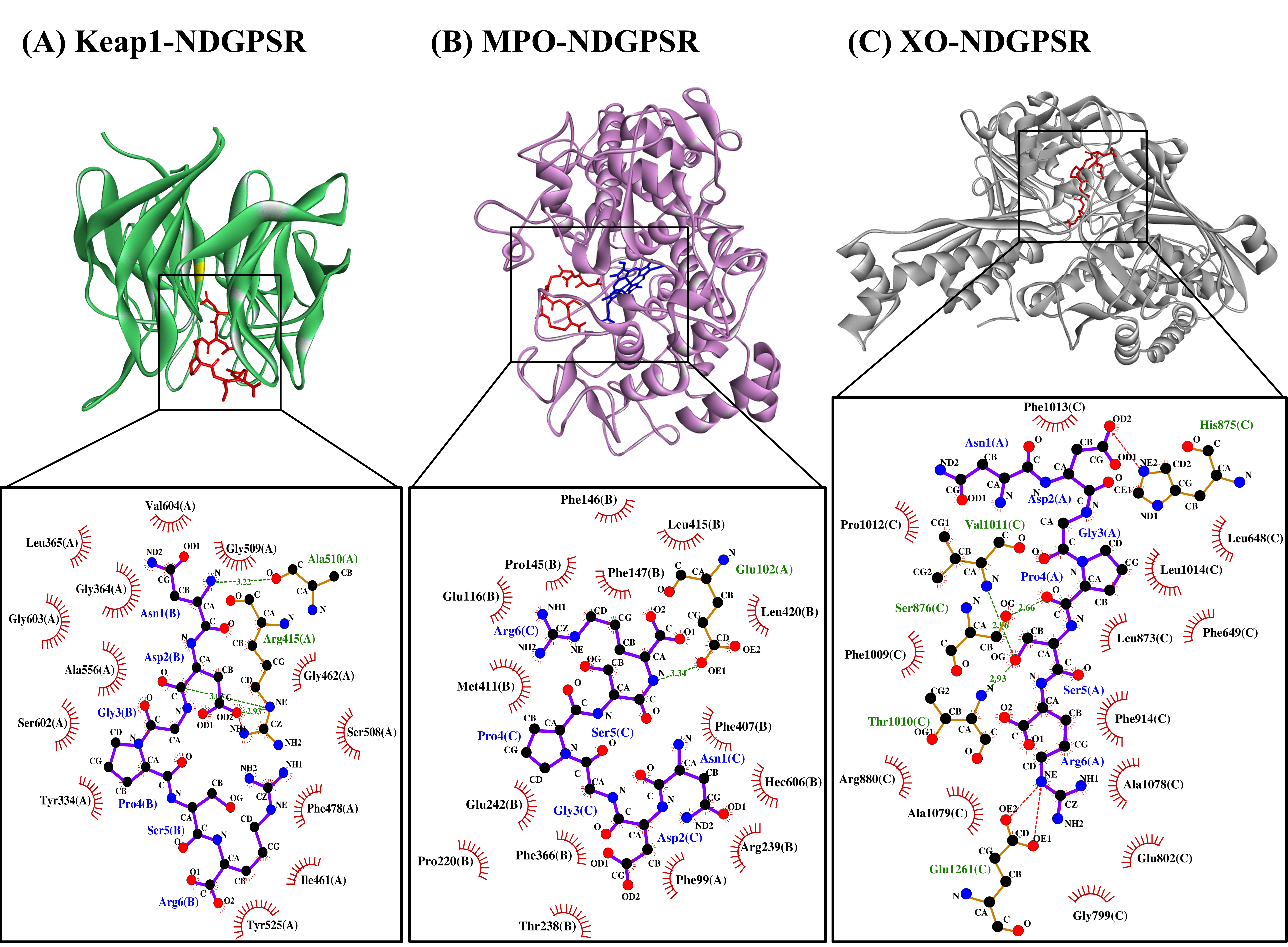
Figure 4. The docked models of NDGPSR interacting with (A) Keap1, (B) MPO, and (C) XO in 3D (top) and 2D (bottom) diagrams. In the 3D diagrams, NDGPSR is displayed in red; the heme moiety of MPO in (B) is displayed in blue. In the 2D diagrams, bonds of proteins are in orange, whereas those of peptides are in purple. Hydrophobic interactions, hydrogen bonds, and salt bridges are represented in red spoked arcs, green dashed lines, and red dashed lines, respectively.
10. Conclusions
In this study, 29 potential antioxidant peptides were purified and identified, for the first time, from a CS hydrolysate. The prevalence of aromatic and basic residues, in addition to binding affinity to ABTS·+, as revealed by molecular docking simulation, may account for the antioxidant activities of the peptides. Our in silico study also unraveled the potential of the peptides as inhibitors of Keap1-Nrf2 interaction, MPO and XO. NDGPSR stood out among the 29 peptides for its concurrent affinities towards the three protein targets, besides being predicted as non-toxic, non-allergenic, and having cell-penetrating potential. Taken together, our findings highlight the potential of CS as a source of antioxidant peptides with desirable properties for future applications in functional food and drug discovery. Future investigations by using in vitro and in vivo models are warranted for more in-depth exploration of CS-derived antioxidant peptides highlighted in this study.

This entry is adapted from the peer-reviewed paper 10.3390/antiox10111822
References
- Hasanudin, K.; Hashim, P.; Mustafa, S. Corn silk (Stigma maydis) in healthcare: A phytochemical and pharmacological review. Molecules 2012, 17, 9697–9715.
- Li, C.-C.; Lee, Y.-C.; Lo, H.-Y.; Huang, Y.-W.; Hsiang, C.-Y.; Ho, T.-Y. Antihypertensive effects of corn silk extract and its novel bioactive constituent in spontaneously hypertensive rats: The involvement of angiotensin-converting enzyme inhibition. Molecules 2019, 24, 1886.
- Ho, T.-Y.; Li, C.-C.; Lo, H.-Y.; Chen, F.-Y.; Hsiang, C.-Y. Corn silk extract and its bioactive peptide ameliorated lipopolysaccharide-induced inflammation in mice via the nuclear factor-κB signaling pathway. J. Agric. Food Chem. 2017, 65, 759–768.
- Chai, T.-T.; Ang, S.-Y.; Goh, K.; Lee, Y.-H.; Ngoo, J.-M.; Teh, L.-K.; Wong, F.-C. Trypsin-hydrolyzed corn silk proteins: Antioxidant activities, in vitro gastrointestinal and thermal stability, and hematoprotective effects. eFood 2020, 1, 156–164.
- Wong, F.-C.; Xiao, J.; Wang, S.; Ee, K.-Y.; Chai, T.-T. Advances on the antioxidant peptides from edible plant sources. Trends Food Sci. Technol. 2020, 99, 44–57.
- Tonolo, F.; Moretto, L.; Grinzato, A.; Fiorese, F.; Folda, A.; Scalcon, V.; Ferro, S.; Arrigoni, G.; Bellamio, M.; Feller, E.; et al. Fermented soy-derived bioactive peptides selected by a molecular docking approach show antioxidant properties involving the Keap1/Nrf2 pathway. Antioxidants 2020, 9, 1306.
- Winkel, A.F.; Engel, C.K.; Margerie, D.; Kannt, A.; Szillat, H.; Glombik, H.; Kallus, C.; Ruf, S.; Güssregen, S.; Riedel, J.; et al. Characterization of RA839, a noncovalent small molecule binder to Keap1 and selective activator of Nrf2 signaling. J. Biol. Chem. 2015, 290, 28446–28455.
- Deng, Z.; Cui, C.; Wang, Y.; Ni, J.; Zheng, L.; Wei, H.-K.; Peng, J. FSGHF3 and peptides, prepared from fish skin gelatin, exert a protective effect on DSS-induced colitis via the Nrf2 pathway. Food Funct. 2020, 11, 414–423.
- Thaha, A.; Wang, B.-S.; Chang, Y.-W.; Hsia, S.-M.; Huang, T.-C.; Shiau, C.-Y.; Hwang, D.-F.; Chen, T.-Y. Food-derived bioactive peptides with antioxidative capacity, xanthine oxidase and tyrosinase inhibitory activity. Processes 2021, 9, 747.
- Sharmila, M.D.; Chai, T.-T.; Wong, F.-C. Antioxidant and protein protection potentials of fennel seed-derived protein hydrolysates and peptides. Mod. Food Sci. Technol. 2019, 35, 22–29.
- Hu, R.; Dunmire, K.M.; Truelock, C.N.; Paulk, C.B.; Aldrich, G.; Li, Y. Antioxidant performances of corn gluten meal and DDGS protein hydrolysates in food, pet food, and feed systems. J. Sci. Food Agric. 2020, 2, 100030.
- Chai, T.-T.; Law, Y.-C.; Wong, F.-C.; Kim, S.-K. Enzyme-assisted discovery of antioxidant peptides from edible marine invertebrates: A review. Mar. Drugs 2017, 15, 42.
- Feng, Y.-X.; Ruan, G.-R.; Jin, F.; Xu, J.; Wang, F.-J. Purification, identification, and synthesis of five novel antioxidant peptides from Chinese chestnut (Castanea mollissima Blume) protein hydrolysates. LWT 2018, 92, 40–46.
- Wang, X.; Zheng, X.; Kopparapu, N.; Cong, W.; Deng, Y.; Sun, X.; Liu, X. Purification and evaluation of a novel antioxidant peptide from corn protein hydrolysate. Process Biochem. 2014, 49, 1562–1569.
- Chai, T.-T.; Xiao, J.; Mohana Dass, S.; Teoh, J.-Y.; Ee, K.-Y.; Ng, W.-J.; Wong, F.-C. Identification of antioxidant peptides derived from tropical jackfruit seed and investigation of the stability profiles. Food Chem. 2021, 340, 127876.
- He, R.; Ju, X.; Yuan, J.; Wang, L.; Girgih, A.T.; Aluko, R.E. Antioxidant activities of rapeseed peptides produced by solid state fermentation. Food Res. Int. 2012, 49, 432–438.
- Osorio, D.; Rondón-Villarreal, P.; Torres, R. Peptides: A package for data mining of antimicrobial peptides. R J 2015, 7, 4–14.
- Wong, F.-C.; Xiao, J.; Ong, M.G.-L.; Pang, M.-J.; Wong, S.-J.; Teh, L.-K.; Chai, T.-T. Identification and characterization of antioxidant peptides from hydrolysate of blue-spotted stingray and their stability against thermal, pH and simulated gastrointestinal digestion treatments. Food Chem. 2019, 271, 614–622.
- Mishra, M.; Nagarajan, K. Assessment of antioxidant influence of short series peptides using hydrogen peroxide scavenging assay and superoxide radical scavenging activity. World J. Pharm. Pharm. Sci. 2018, 7, 1064–1070.
- Chen, T.; Chen, Z.; Wang, H.; Chen, X.; Yang, J.; Han, A.; Lin, D.-H.; Hong, J. Underlying action mechanism of a novel antioxidant peptide derived from Allium tuberosum Rottler protein hydrolysates and its protective effects on hydrogen peroxide induced cell injury. J. Funct. Foods 2018, 40, 606–613.
- Olsen, T.H.; Yesiltas, B.; Marin, F.I.; Pertseva, M.; García-Moreno, P.J.; Gregersen, S.; Overgaard, M.T.; Jacobsen, C.; Lund, O.; Hansen, E.B.; et al. AnOxPePred: Using deep learning for the prediction of antioxidative properties of peptides. Sci. Rep. 2020, 10, 21471.
- Kovacs-Nolan, J.; Zhang, H.; Ibuki, M.; Nakamori, T.; Yoshiura, K.; Turner, P.V.; Matsui, T.; Mine, Y. The PepT1-transportable soy tripeptide VPY reduces intestinal inflammation. Biochim. Biophys. Acta 2012, 1820, 1753–1763.
- He, S.; Zhang, Y.; Sun, H.; Du, M.; Qiu, J.; Tang, M.; Sun, X.; Zhu, B. Antioxidative peptides from proteolytic hydrolysates of false abalone (Volutharpa ampullacea perryi): Characterization, identification, and molecular docking. Mar. Drugs 2019, 17, 116.
- Shaw, S.A.; Vokits, B.P.; Dilger, A.K.; Viet, A.; Clark, C.G.; Abell, L.M.; Locke, G.A.; Duke, G.; Kopcho, L.M.; Dongre, A.; et al. Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase. Bioorg. Med. Chem. 2020, 28, 115723.
- Zhang, Y.; He, S.; Bonneil, É.; Simpson, B.K. Generation of antioxidative peptides from Atlantic sea cucumber using alcalase versus trypsin: In vitro activity, de novo sequencing, and in silico docking for in vivo function prediction. Food Chem. 2020, 306, 125581.
- Zhong, H.; Abdullah; Zhang, Y.; Deng, L.; Zhao, M.; Tang, J.; Zhang, H.; Feng, F.; Wang, J. Exploring the potential of novel xanthine oxidase inhibitory peptide (ACECD) derived from Skipjack tuna hydrolysates using affinity-ultrafiltration coupled with HPLC-MALDI-TOF/TOF-MS. Food Chem. 2021, 347, 129068.
- Cao, H.; Pauff, J.M.; Hille, R. X-ray crystal structure of a xanthine oxidase complex with the flavonoid inhibitor quercetin. J. Nat. Prod. 2014, 77, 1693–1699.
- Thaha, A.; Wang, B.-S.; Chang, Y.-W.; Hsia, S.-M.; Huang, T.-C.; Shiau, C.-Y.; Hwang, D.-F.; Chen, T.-Y. Food-derived bioactive peptides with antioxidative capacity, xanthine oxidase and tyrosinase inhibitory activity. Processes 2021, 9, 747.
- Chai, T.-T.; Law, Y.-C.; Wong, F.-C.; Kim, S.-K. Enzyme-assisted discovery of antioxidant peptides from edible marine invertebrates: A review. Mar. Drugs 2017, 15, 42.