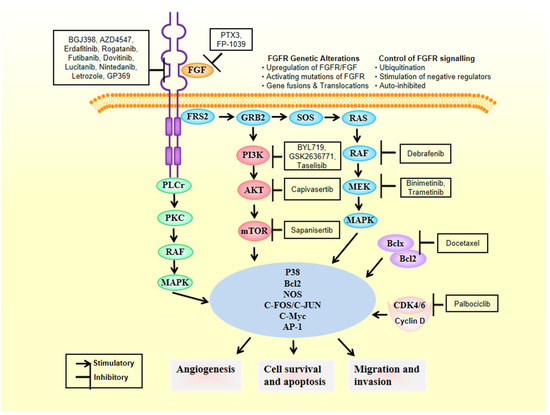
2. FGFRs as Oncogenic Drivers
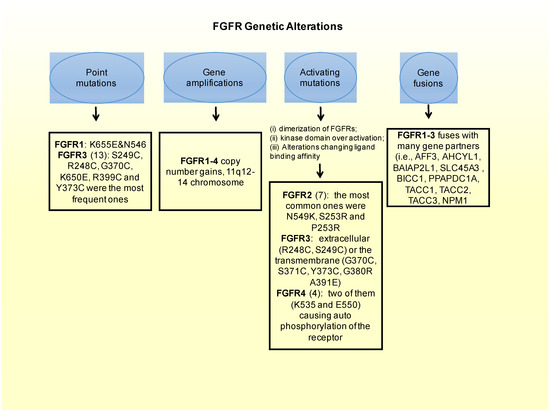
3. FGFR Genetic Alterations in Breast Cancer
3.1. FGFRs Gene Amplification
3.2. FGFRs Activating Mutations
-
In FGFR2: there are 12 mutations reported in a Catalog of Somatic Mutations in Cancer (COSMIC), which is the largest database entailing breast cancer somatic mutations. There are only seven missense mutations capable of constitutively activate the receptor. Among these, the most common ones in FGFR2 are N549K, S253R and P253R [43]. Moreover these three activating mutations are located on the extracellular region of the receptors between the two immunoglobulin-like domains, domains that are important for ligand binding [63]. In estrogen receptor positive breast cancers the M538I and N550K mutations of FGFR2 contribute to giving resistance to inhibitors of SERDs and CDK4/6. Moreover, in some cohorts of estrogen receptor positive MBC patients resistant to CDK4/6 and SERDs FGFR2 mutations were detected. This could imply that FGFR2 could be involved in a mechanism conferring some resistance to patients. Therefore FGFR2 mutated patients could benefit most from the combination of CDK4/6, SERDs and FGFR inhibitors.
-
FGFR3: from the COSMIC database, 13 point-mutations were detected. Among them, S249C, R248C, G370C, K650E, R399C and Y373C were the most frequent ones. Frequent activating mutations in this gene affect either the extracellular (R248C, S249C) or the transmembrane (G370C, S371C, Y373C, G380R A391E) protein domains. There are also a number of rare mutations within the kinase domain, such as, K650E, K650N, K650M, K650T K650Q, and N540S [43,64].
3.3. Fusion of FGFRs Genes
3.4. Genome-Wide Studies
4. Anti-FGFR Therapies
5. Discussion and Conclusions
| BC | Breast Cancer |
| FGF | Fibroblast Growth Factor |
| FGFR | Fibroblast Growth Factor Receptor |
| VEGFR | Vascular Endothelial Growth Factor |
| HPSGs | heparan sulfate proteoglycans |
| EC | N-terminal extracellular |
| TM | transmembrane |
| IC | intracellular |
| PI3K | phosphoinositide-3-kinase |
| MAPK | mitogen-activated protein kinase |
| 4-OHT | 4-hydroxytamoxifen |
| TICs | maintaining tumor-initiating cells |
| GWAS | Genome-Wide-Association-Studies |
| PKC | activates protein kinase C |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Sobhani, N.; Ianza, A.; D’Angelo, A.; Roviello, G.; Giudici, F.; Bortul, M.; Zanconati, F.; Bottin, C.; Generali, D. Current Status of Fibroblast Growth Factor Receptor-Targeted Therapies in Breast Cancer. Cells 2018, 7, 76. [Google Scholar] [CrossRef] [PubMed]
- Dienstmann, R.; Rodon, J.; Prat, A.; Perez-Garcia, J.; Adamo, B.; Felip, E.; Cortes, J.; Iafrate, A.J.; Nuciforo, P.; Tabernero, J. Genomic aberrations in the FGFR pathway: Opportunities for targeted therapies in solid tumors. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2014, 25, 552–563. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.; Wynshaw-Boris, A.; Zhou, F.; Kuo, A.; Leder, P. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell 1996, 84, 911–921. [Google Scholar] [CrossRef]
- Presta, M.; Chiodelli, P.; Giacomini, A.; Rusnati, M.; Ronca, R. Fibroblast growth factors (FGFs) in cancer: FGF traps as a new therapeutic approach. Pharmacol. Ther. 2017, 179, 171–187. [Google Scholar] [CrossRef]
- Belov, A.A.; Mohammadi, M. Molecular mechanisms of fibroblast growth factor signaling in physiology and pathology. Cold Spring Harb. Perspect. Biol. 2013, 5, a015958. [Google Scholar] [CrossRef]
- Babina, I.S.; Turner, N.C. Advances and challenges in targeting FGFR signalling in cancer. Nat. Rev. Cancer 2017, 17, 318–332. [Google Scholar] [CrossRef]
- Ornitz, D.M.; Itoh, N. Fibroblast growth factors. Genome Biol. 2001, 2, REVIEWS3005. [Google Scholar] [CrossRef]
- Eswarakumar, V.P.; Lax, I.; Schlessinger, J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005, 16, 139–149. [Google Scholar] [CrossRef]
- Porta, R.; Borea, R.; Coelho, A.; Khan, S.; Araújo, A.; Reclusa, P.; Franchina, T.; van der Steen, N.; van Dam, P.; Ferri, J.; et al. FGFR a promising druggable target in cancer: Molecular biology and new drugs. Crit. Rev. Oncol. 2017, 113, 256–267. [Google Scholar] [CrossRef]
- André, F.; Cortés, J. Rationale for targeting fibroblast growth factor receptor signaling in breast cancer. Breast Cancer Res. Treat. 2015, 150, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.; Grose, R. Fibroblast growth factor signalling: From development to cancer. Nat. Rev. Cancer 2010, 10, 116–129. [Google Scholar] [CrossRef]
- Ornitz, D.M.; Xu, J.; Colvin, J.S.; McEwen, D.G.; MacArthur, C.A.; Coulier, F.; Gao, G.; Goldfarb, M. Receptor specificity of the fibroblast growth factor family. J. Biol. Chem. 1996, 271, 15292–15297. [Google Scholar] [CrossRef] [PubMed]
- Tanner, Y.; Grose, R.P. Dysregulated FGF signalling in neoplastic disorders. Semin. Cell Dev. Biol. 2016, 53, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Katoh, M.; Nakagama, H. FGF Receptors: Cancer Biology and Therapeutics. Med. Res. Rev. 2014, 34, 280–300. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, M.; Dikic, I.; Sorokin, A.; Burgess, W.H.; Jaye, M.; Schlessinger, J. Identification of six novel autophosphorylation sites on fibroblast growth factor receptor 1 and elucidation of their importance in receptor activation and signal transduction. Mol. Cell. Biol. 1996, 16, 977–989. [Google Scholar] [CrossRef]
- Yashiro, M.; Matsuoka, T. Fibroblast growth factor receptor signaling as therapeutic targets in gastric cancer. World J. Gastroenterol. 2016, 22, 2415. [Google Scholar] [CrossRef]
- Chell, V.; Balmanno, K.; Little, A.S.; Wilson, M.; Andrews, S.; Blockley, L. Tumour cell responses to new fibroblast growth factor receptor tyrosine kinase inhibitors and identification of a gatekeeper mutation in FGFR3 as a mechanism of acquired resistance. Oncogene 2013, 32, 3059–3070. [Google Scholar] [CrossRef]
- Tiong, K.H.; Mah, L.Y.; Leong, C.-O. Functional roles of fibroblast growth factor receptors (FGFRs) signaling in human cancers. Apoptosis 2013, 18, 1447–1468. [Google Scholar] [CrossRef]
- Katoh, M. FGFR inhibitors: Effects on cancer cells, tumor microenvironment and whole-body homeostasis (Review). Int. J. Mol. Med. 2016, 38, 3–15. [Google Scholar] [CrossRef]
- Li, X.; Wang, C.; Xiao, J.; McKeehan, W.L.; Wang, F. Fibroblast growth factors, old kids on the new block. Semin. Cell Dev. Biol. 2016, 53, 155–167. [Google Scholar] [CrossRef] [PubMed]
- Hallinan, N.; Finn, S.; Cuffe, S.; Rafee, S.; O’Byrne, K.; Gately, K. Targeting the fibroblast growth factor receptor family in cancer. Cancer Treat. Rev. 2016, 46, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Rudert, W.A.; Loutaev, I.; Roginskaya, V.; Corey, S.J. Repression of c-Cbl leads to enhanced G-CSF Jak-STAT signaling without increased cell proliferation. Oncogene 2002, 21, 5346–5355. [Google Scholar] [CrossRef] [PubMed]
- Kovalenko, D.; Yang, X.; Nadeau, R.J.; Harkins, L.K.; Friesel, R. Sef inhibits fibroblast growth factor signaling by inhibiting FGFR1 tyrosine phosphorylation and subsequent ERK activation. J. Biol. Chem. 2003, 278, 14087–14091. [Google Scholar] [CrossRef]
- Torii, S.; Kusakabe, M.; Yamamoto, T.; Maekawa, M.; Nishida, E. Sef Is a Spatial Regulator for Ras/MAP Kinase Signaling. Dev. Cell 2004, 7, 33–44. [Google Scholar] [CrossRef]
- Goetz, R.; Mohammadi, M. Exploring mechanisms of FGF signalling through the lens of structural biology. Nat. Rev. Mol. Cell Biol. 2013, 14, 166–180. [Google Scholar] [CrossRef]
- Stauber, D.J.; DiGabriele, A.D.; Hendrickson, W.A. Structural interactions of fibroblast growth factor receptor with its ligands. Proc. Natl. Acad. Sci. USA 2000, 97, 49–54. [Google Scholar] [CrossRef]
- Herbert, C.; Lassalle, G.; Alcouffe, C.; Bono, F. Approaches targeting the FGF–FGFR system: A review of the recent patent literature and associated advanced therapeutic agents. Pharm. Pat. Anal. 2014, 3, 585–612. [Google Scholar] [CrossRef]
- Fearon, A.E.; Gould, C.R.; Grose, R.P. FGFR signalling in women’s cancers. Int. J. Biochem. Cell Biol. 2013, 45, 2832–2842. [Google Scholar] [CrossRef]
- Lasorella, A.; Sanson, M.; Iavarone, A. FGFR-TACC gene fusions in human glioma. Neuro Oncol. 2016, 19, 475–483. [Google Scholar] [CrossRef]
- Touat, M.; Ileana, E.; Postel-Vinay, S.; Andre, F.; Soria, J.-C. Targeting FGFR Signaling in Cancer. Clin. Cancer Res. 2015, 21, 2684–2694. [Google Scholar] [CrossRef] [PubMed]
- Saichaemchan, S.; Ariyawutyakorn, W.; Varella-Garcia, M. Fibroblast Growth Factor Receptors: From the Oncogenic Pathway to Targeted Therapy. Curr. Mol. Med. 2016, 16, 40–62. [Google Scholar] [CrossRef] [PubMed]
- Katoh, M. Therapeutics Targeting FGF Signaling Network in Human Diseases. Trends Pharmacol. Sci. 2016, 37, 1081–1096. [Google Scholar] [CrossRef] [PubMed]
- Sarabipour, S.; Hristova, K. Mechanism of FGF receptor dimerization and activation. Nat. Commun. 2016, 7, 10262. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, Y. Therapeutic uses of FGFs. Semin. Cell Dev. Biol. 2016, 53, 144–154. [Google Scholar] [CrossRef]
- Seo, A.N.; Lee, H.J.; Kim, E.J.; Kim, H.J.; Jang, M.H.; Lee, H.E.; Kim, Y.J.; Kim, J.H.; Park, S.Y. Tumour-infiltrating CD8+ lymphocytes as an independent predictive factor for pathological complete response to primary systemic therapy in breast cancer. Br. J. Cancer 2013, 109, 2705–2713. [Google Scholar] [CrossRef]
- Xie, L.; Su, X.; Zhang, L.; Yin, X.; Tang, L.; Zhang, X.; Xu, Y.; Gao, Z.; Liu, K.; Zhou, M.; et al. FGFR2 gene amplification in gastric cancer predicts sensitivity to the selective FGFR inhibitor AZD4547. Clin. Cancer Res. 2013, 19, 2572–2583. [Google Scholar] [CrossRef]
- Turner, N.; Pearson, A.; Sharpe, R.; Lambros, M.; Geyer, F.; Lopez-Garcia, M.A.; Natrajan, R.; Marchio, C.; Iorns, E.; Mackay, A.; et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 2010, 70, 2085–2094. [Google Scholar] [CrossRef]
- Ronca, R.; Giacomini, A.; Rusnati, M.; Presta, M. The potential of fibroblast growth factor/fibroblast growth factor receptor signaling as a therapeutic target in tumor angiogenesis. Expert Opin. Ther. Targets 2015, 19, 1361–1377. [Google Scholar] [CrossRef]
- Kumar, S.B.V.S.; Narasu, L.; Gundla, R.; Dayam, R.; Sarma, J.A.R.P. Fibroblast growth factor receptor inhibitors. Curr. Pharm. Des. 2013, 19, 687–701. [Google Scholar] [CrossRef]
- Kelleher, F.C.; O’Sullivan, H.; Smyth, E.; McDermott, R.; Viterbo, A. Fibroblast growth factor receptors, developmental corruption and malignant disease. Carcinogenesis 2013, 34, 2198–2205. [Google Scholar] [CrossRef] [PubMed]
- Helsten, T.; Schwaederle, M.; Kurzrock, R. Fibroblast growth factor receptor signaling in hereditary and neoplastic disease: Biologic and clinical implications. Cancer Metastasis Rev. 2015, 34, 479–496. [Google Scholar] [CrossRef] [PubMed]
- Helsten, T.; Elkin, S.; Arthur, E.; Tomson, B.N.; Carter, J.; Kurzrock, R. The FGFR Landscape in Cancer: Analysis of 4,853 Tumors by Next-Generation Sequencing. Clin. Cancer Res. 2016, 22, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Carter, E.P.; Fearon, A.E.; Grose, R.P. Careless talk costs lives: Fibroblast growth factor receptor signalling and the consequences of pathway malfunction. Trends Cell Biol. 2015, 25, 221–233. [Google Scholar] [CrossRef]
- Wesche, J.; Haglund, K.; Haugsten, E.M. Fibroblast growth factors and their receptors in cancer. Biochem. J. 2011, 437, 199–213. [Google Scholar] [CrossRef]
- Heinzle, C.; Sutterlüty, H.; Grusch, M.; Grasl-Kraupp, B.; Berger, W.; Marian, B. Targeting fibroblast-growth-factor-receptor-dependent signaling for cancer therapy. Expert Opin. Ther. Targets 2011, 15, 829–846. [Google Scholar] [CrossRef]
- Greulich, H.; Pollock, P.M. Targeting mutant fibroblast growth factor receptors in cancer. Trends Mol. Med. 2011, 17, 283–292. [Google Scholar] [CrossRef]
- Adnane, J.; Gaudray, P.; Dionne, C.A.; Crumley, G.; Jaye, M.; Schlessinger, J.; Jeanteur, P.; Birnbaum, D.; Theillet, C. BEK and FLG, two receptors to members of the FGF family, are amplified in subsets of human breast cancers. Oncogene 1991, 6, 659–663. [Google Scholar]
- Dey, N.; Williams, C.; Leyland-Jones, B.; De, P. Mutation matters in precision medicine: A future to believe in. Cancer Treat. Rev. 2017, 55, 136–149. [Google Scholar] [CrossRef]
- Shao, F.; Sun, H.; Deng, C.-X. Potential therapeutic targets of triple-negative breast cancer based on its intrinsic subtype. Oncotarget 2017, 8, 73329–73344. [Google Scholar] [CrossRef]
- Elbauomy Elsheikh, S.; Green, A.R.; Lambros, M.B.; Turner, N.C.; Grainge, M.J.; Powe, D.; Ellis, I.O.; Reis-Filho, J.S. FGFR1 amplification in breast carcinomas: A chromogenic in situ hybridisation analysis. Breast Cancer Res. 2007, 9, R23. [Google Scholar] [CrossRef] [PubMed]
- Massard, C.; Michiels, S.; Ferté, C.; Le Deley, M.-C.; Lacroix, L.; Hollebecque, A.; Verlingue, L.; Ileana, E.; Rosellini, S.; Ammari, S.; et al. High-Throughput Genomics and Clinical Outcome in Hard-to-Treat Advanced Cancers: Results of the MOSCATO 01 Trial. Cancer Discov. 2017, 7, 586–595. [Google Scholar] [CrossRef] [PubMed]
- André, F.; Bachelot, T.; Commo, F.; Campone, M.; Arnedos, M.; Dieras, V.; Lacroix-Triki, M.; Lacroix, L.; Cohen, P.; Gentien, D.; et al. Comparative genomic hybridisation array and DNA sequencing to direct treatment of metastatic breast cancer: A multicentre, prospective trial (SAFIR01/UNICANCER). Lancet Oncol. 2014, 15, 267–274. [Google Scholar] [CrossRef]
- Brunello, E.; Brunelli, M.; Bogina, G.; Caliò, A.; Manfrin, E.; Nottegar, A.; Vergine, M.; Molino, A.; Bria, E.; Massari, F.; et al. FGFR-1 amplification in metastatic lymph-nodal and haematogenous lobular breast carcinoma. J. Exp. Clin. Cancer Res. 2012, 31, 103. [Google Scholar] [CrossRef]
- Reis-Filho, J.S.; Simpson, P.T.; Turner, N.C.; Lambros, M.B.; Jones, C.; Mackay, A.; Grigoriadis, A.; Sarrio, D.; Savage, K.; Dexter, T.; et al. FGFR1 Emerges as a Potential Therapeutic Target for Lobular Breast Carcinomas. Clin. Cancer Res. 2006, 12, 6652–6662. [Google Scholar] [CrossRef]
- Courjal, F.; Cuny, M.; Simony-Lafontaine, J.; Louason, G.; Speiser, P.; Zeillinger, R.; Rodriguez, C.; Theillet, C. Mapping of DNA amplifications at 15 chromosomal localizations in 1875 breast tumors: Definition of phenotypic groups. Cancer Res. 1997, 57, 4360–4367. [Google Scholar]
- Andre, F.; Bachelot, T.; Campone, M.; Dalenc, F.; Perez-Garcia, J.M.; Hurvitz, S.A.; Turner, N.; Rugo, H.; Smith, J.W.; Deudon, S.; et al. Targeting FGFR with Dovitinib (TKI258): Preclinical and Clinical Data in Breast Cancer. Clin. Cancer Res. 2013, 19, 3693–3702. [Google Scholar] [CrossRef]
- Formisano, L.; Lu, Y.; Servetto, A.; Hanker, A.B.; Jansen, V.M.; Bauer, J.A.; Sudhan, D.R.; Guerrero-Zotano, A.L.; Croessmann, S.; Guo, Y.; et al. Aberrant FGFR signaling mediates resistance to CDK4/6 inhibitors in ER+ breast cancer. Nat. Commun. 2019, 10, 1373. [Google Scholar] [CrossRef]
- Kim, S.; Dubrovska, A.; Salamone, R.J.; Walker, J.R.; Grandinetti, K.B.; Bonamy, G.M.C.; Orth, A.P.; Elliott, J.; Porta, D.G.; Garcia-Echeverria, C.; et al. FGFR2 promotes breast tumorigenicity through maintenance of breast tumor-initiating cells. PLoS ONE 2013, 8, e51671. [Google Scholar] [CrossRef]
- Guagnano, V.; Kauffmann, A.; Wohrle, S.; Stamm, C.; Ito, M.; Barys, L.; Pornon, A.; Yao, Y.; Li, F.; Zhang, Y.; et al. FGFR Genetic Alterations Predict for Sensitivity to NVP-BGJ398, a Selective Pan-FGFR Inhibitor. Cancer Discov. 2012, 2, 1118–1133. [Google Scholar] [CrossRef]
- Hart, K.C.; Robertson, S.C.; Kanemitsu, M.Y.; Meyer, A.N.; Tynan, J.A.; Donoghue, D.J. Transformation and Stat activation by derivatives of FGFR1, FGFR3 and FGFR4. Oncogene 2000, 19, 3309–3320. [Google Scholar] [CrossRef] [PubMed]
- Lew, E.D.; Furdui, C.M.; Anderson, K.S.; Schlessinger, J. The Precise Sequence of FGF Receptor Autophosphorylation Is Kinetically Driven and Is Disrupted by Oncogenic Mutations. Sci. Signal. 2009, 2, ra6. [Google Scholar] [CrossRef] [PubMed]
- Dutt, A.; Salvesen, H.B.; Chen, T.H.; Ramos, A.H.; Onofrio, R.C.; Hatton, C.; Nicoletti, R.; Winckler, W.; Grewal, R.; Hanna, M.; et al. Drug-sensitive FGFR2 mutations in endometrial carcinoma. Proc. Natl. Acad. Sci. USA 2008, 105, 8713–8717. [Google Scholar] [CrossRef] [PubMed]
- Pandith, A.A.; Shah, Z.A.; Siddiqi, M.A. Oncogenic role of fibroblast growth factor receptor 3 in tumorigenesis of urinary bladder cancer. Urol. Oncol. Semin. Orig. Investig. 2013, 31, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Guo, W.; Shen, J.K.; Mankin, H.J.; Hornicek, F.J.; Duan, Z. Rhabdomyosarcoma: Advances in Molecular and Cellular Biology. Sarcoma 2015, 2015, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Parker, B.C.; Engels, M.; Annala, M.; Zhang, W. Emergence of FGFR family gene fusions as therapeutic targets in a wide spectrum of solid tumours. J. Pathol. 2014, 232, 4–15. [Google Scholar] [CrossRef]
- Nelson, K.N.; Meyer, A.N.; Siari, A.; Campos, A.R.; Motamedchaboki, K.; Donoghue, D.J. Oncogenic Gene Fusion FGFR3-TACC3 Is Regulated by Tyrosine Phosphorylation. Mol. Cancer Res. 2016, 14, 458–469. [Google Scholar] [CrossRef]
- Wu, Y.-M.; Su, F.; Kalyana-Sundaram, S.; Khazanov, N.; Ateeq, B.; Cao, X.; Lonigro, R.J.; Vats, P.; Wang, R.; Lin, S.F.; et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013, 3, 636–647. [Google Scholar] [CrossRef]
- Stransky, N.; Cerami, E.; Schalm, S.; Kim, J.L.; Lengauer, C. The landscape of kinase fusions in cancer. Nat. Commun. 2014, 5, 4846. [Google Scholar] [CrossRef]
- Easton, D.F.; Pooley, K.A.; Dunning, A.M.; Pharoah, P.D.P.; Thompson, D.; Ballinger, D.G.; Struewing, J.P.; Morrison, J.; Field, H.; Luben, R.; et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 2007, 447, 1087–1093. [Google Scholar] [CrossRef]
- Stacey, S.N.; Manolescu, A.; Sulem, P.; Thorlacius, S.; Gudjonsson, S.A.; Jonsson, G.F.; Jakobsdottir, M.; Bergthorsson, J.T.; Gudmundsson, J.; Aben, K.K.; et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor–positive breast cancer. Nat. Genet. 2008, 40, 703–706. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.B.; Maia, A.-T.; O’Reilly, M.; Teschendorff, A.E.; Chin, S.; Caldas, C.; Ponder, B.A.J. Allele-specific up-regulation of FGFR2 increases susceptibility to breast cancer. PLoS Biol. 2008, 6, e108. [Google Scholar] [CrossRef] [PubMed]
- Hunter, D.J.; Kraft, P.; Jacobs, K.B.; Cox, D.G.; Yeager, M.; Hankinson, S.E.; Wacholder, S.; Wang, Z.; Welch, R.; Hutchinson, A.; et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007, 39, 870–874. [Google Scholar] [CrossRef] [PubMed]
- Penault-Llorca, F.; Bertucci, F.; Adélaïde, J.; Parc, P.; Coulier, F.; Jacquemier, J.; Birnbaum, D.; Delapeyrière, O. Expression of FGF and FGF receptor genes in human breast cancer. Int. J. Cancer 1995, 61, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Bedussi, F.; Bottini, A.; Memo, M.; Fox, S.B.; Sigala, S.; Generali, D. Targeting fibroblast growth factor receptor in breast cancer: A promise or a pitfall? Expert Opin. Ther. Targets 2014, 18, 665–678. [Google Scholar] [CrossRef]
- Sahores, A.; May, M.; Sequeira, G.; Fuentes, C.; Jacobsen, B.; Lanari, C.; Lamb, C.A. Targeting FGFR with BGJ398 in breast cancer: Effect on tumor growth and metastasis. Curr. Cancer Drug Targets 2017, 18, 979–987. [Google Scholar] [CrossRef]
- Dizman, N.; Rosenberg, J.E.; Hoffman-Censits, J.H.; Quinn, D.I.; Petrylak, D.; Galsky, M.; Vaishampayan, U.; De Giorgi, U.; Gupta, S.; Howard Burris, H.; et al. Infigratinib in upper tract urothelial carcinoma vs urothelial carcinoma of the bladder and association with comprehensive genomic profiling/cell-free DNA results. J. Clin. Oncol. 2019, 37, 4510. [Google Scholar] [CrossRef]
- Kalinina, J.; Dutta, K.; Ilghari, D.; Beenken, A.; Goetz, R.; Eliseenkova, A.V.; Cowburn, D.; Mohammadi, M. The Alternatively Spliced Acid Box Region Plays a Key Role in FGF Receptor Autoinhibition. Structure 2012, 20, 77–88. [Google Scholar] [CrossRef]
- De Luca, A.; Frezzetti, D.; Gallo, M.; Normanno, N. FGFR-targeted therapeutics for the treatment of breast cancer. Expert Opin. Investig. Drugs 2017, 26, 303–311. [Google Scholar] [CrossRef]
- Bai, A.; Meetze, K.; Vo, N.Y.; Kollipara, S.; Mazsa, E.K.; Winston, W.M.; Weiler, S.; Poling, L.L.; Chen, T.; Ismail, N.S.; et al. GP369, an FGFR2-IIIb-Specific Antibody, Exhibits Potent Antitumor Activity against Human Cancers Driven by Activated FGFR2 Signaling. Cancer Res. 2010, 70, 7630–7639. [Google Scholar] [CrossRef]
- Sharpe, R.; Pearson, A.; Herrera-Abreu, M.T.; Johnson, D.; Mackay, A.; Welti, J.C.; Natrajan, R.; Reynolds, A.R.; Reis-Filho, J.S.; Ashworth, A.; et al. FGFR signaling promotes the growth of triple-negative and basal-like breast cancer cell lines both In Vitro and In Vivo. Clin. Cancer Res. 2011, 17, 5275–5286. [Google Scholar] [CrossRef] [PubMed]
- Tolcher, A.W.; Papadopoulos, K.P.; Patnaik, A.; Wilson, K.; Thayer, S.; Zanghi, J.; Gemo, A.T.; Kavanaugh, W.M.; Keer, H.N.; LoRusso, P.M. A phase I, first in human study of FP-1039 (GSK3052230), a novel FGF ligand trap, in patients with advanced solid tumors. Ann. Oncol. 2016, 27, 526–532. [Google Scholar] [CrossRef] [PubMed]