Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Adaptive evolution is a process in which variation that confers an evolutionary advantage in a specific environmental context arises and is propagated through a population. When investigating this process, researchers have mainly focused on describing advantageous phenotypes or putative advantageous genotypes. The main molecular mechanism underlying adaptive evolution in vertebrates is mutations, leading to regulatory changes in gene expression and/or cellular pathways.
- adaptive evolution
- vertebrates
- regulatory mechanisms
1. Introduction
Vertebrates colonized a broad range of habitats with varying ecological conditions (e.g., lighting conditions, oxygen content, and water salinity). This process was associated with adaptations to these varied environments. Adaptive evolution is a process by which advantageous phenotypes, i.e., traits which increase fitness, are propagated through positive selection or environmentally induced variation [1]. Advantageous traits are determined by evolutionary pressures and habitat dynamics; consequently, a phenotype that is advantageous in certain ecological contexts might be neutral or deleterious in others.
Adaptive evolutionary processes occur in two general steps: (1) variation arises and (2) advantageous phenotype is propagated and maintained in the population. Genetic variation arises from mutations that can occur due to single base changes, insertions, deletions, and duplications [2]. Mutations can have multiple implications, from pseudogenization to de novo emergence of genes and/or regulatory elements (Figure 1). Mutations can give rise to novel phenotypes, and the advantageous ones become propagated in a population [2]. Thus, functionally important genomic regions are assumed to be highly conserved and adaptive alleles undergo rapid fixation [3][4]. The conserved nature of these functionally important elements can be used to identify them as being adaptive.
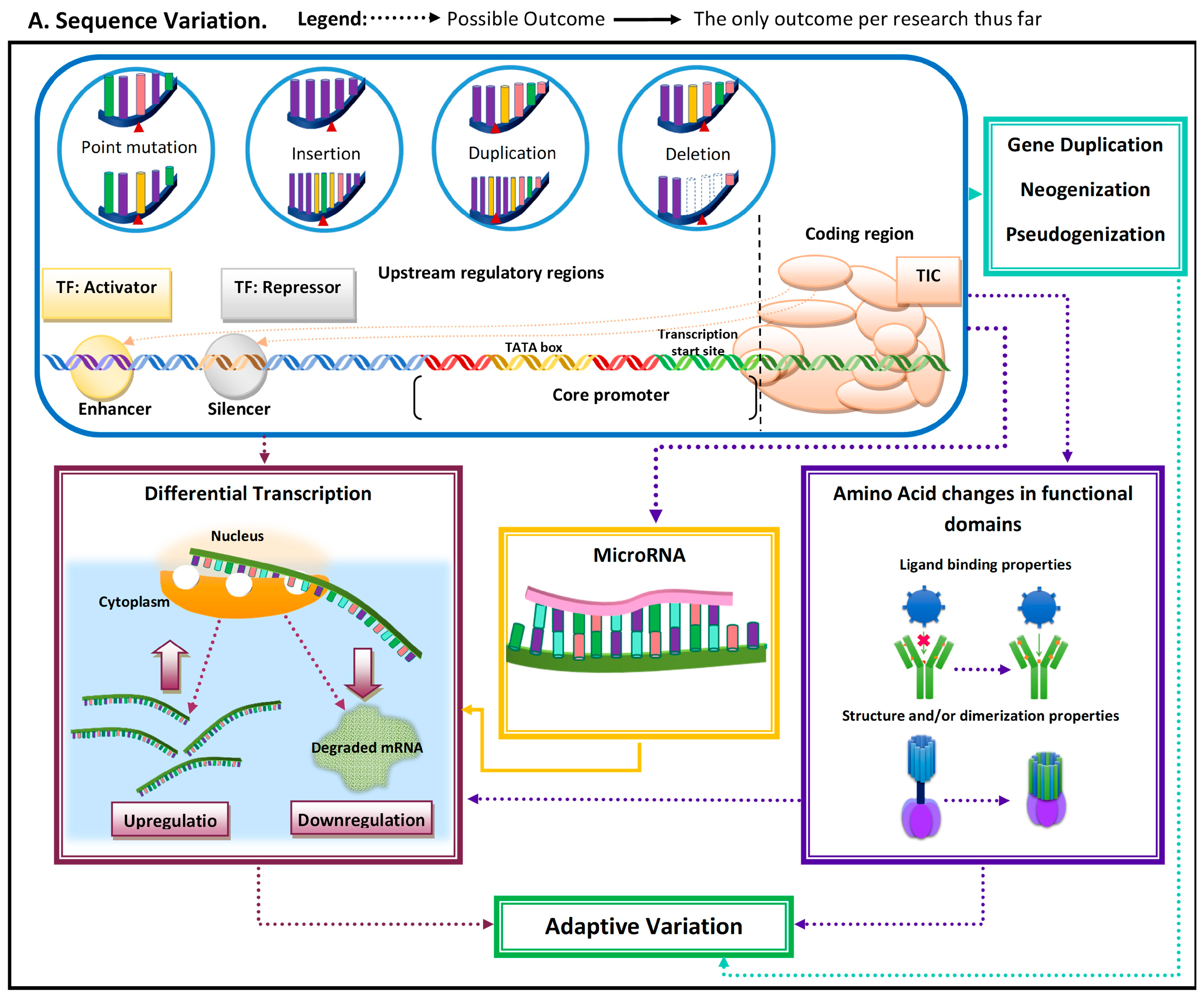
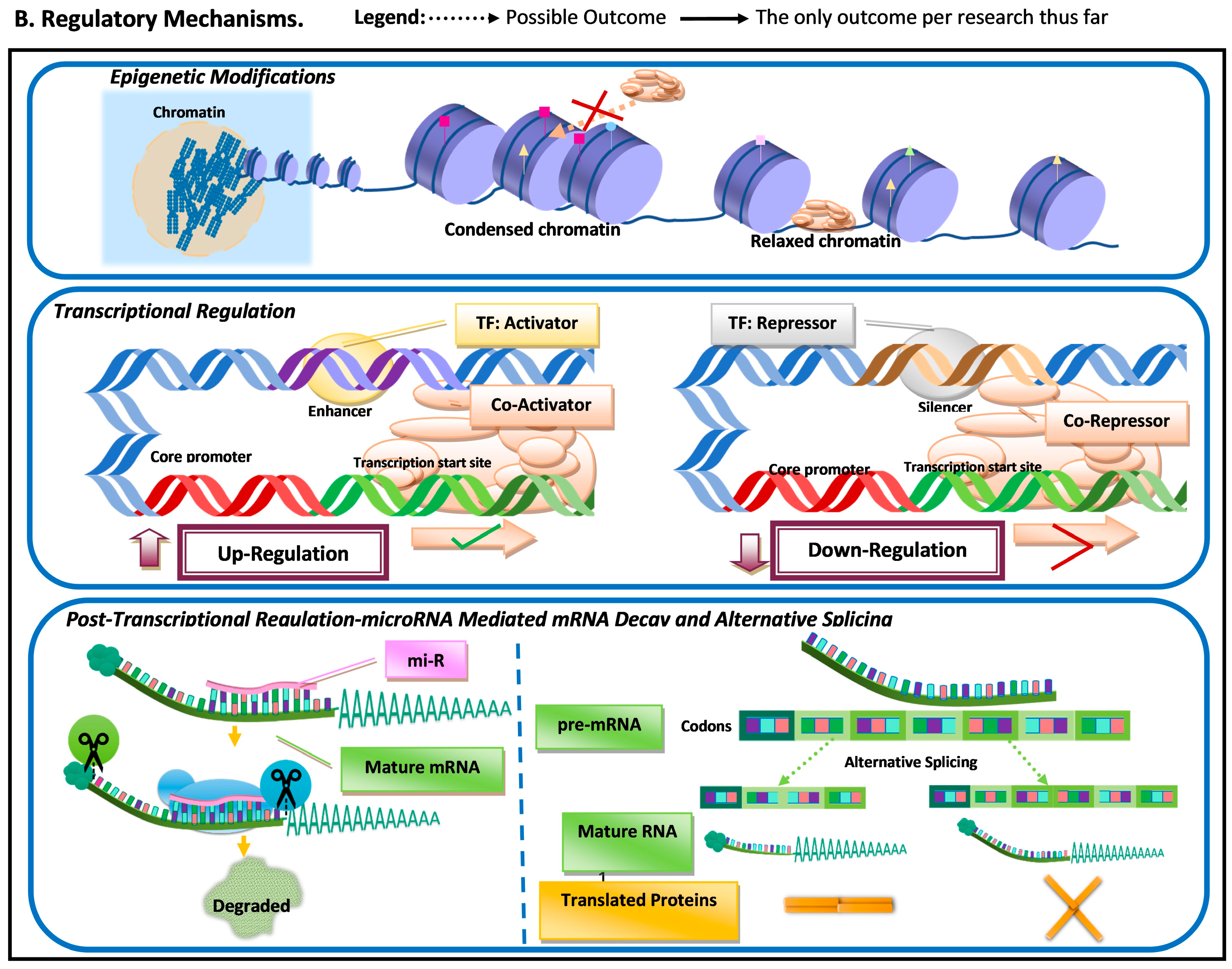
Figure 1. Schematic representation of the most common molecular mechanisms underlying adaptive variation in vertebrates. (A) Sequence variation. Sequence variation refers to any change in DNA sequence that commonly occurs due to DNA polymerase errors (e.g., slippage and erroneous excisions) and/or insertion or deletion of transposable elements. DNA sequence variation in conserved elements (i.e., regulatory elements and coding regions) is the most common mechanism underlying adaptive variation in vertebrates and can lead to gene duplication, neogenization, and pseudogenization. Sequence variation in coding regions can lead to amino acid changes at or near functional domains, which can impact protein function and/or structure. Additionally, sequence variation at the coding regions can also lead to the emergence of microRNA (miR) or modification in existing miRs. MiRs can hybridize with target mRNA and can lead to its decay, thus downregulating gene expression. Sequence variation in regulatory elements can lead to increases (upregulation) or decreases (downregulation) in transcription of a gene. (B). Regulatory mechanisms. The three most common regulatory mechanisms underlying adaptive variation in vertebrates described thus far are epigenetic modifications, transcriptional regulation, and post-transcriptional regulation. Epigenetic modifications are modifications to histones, the protein component of nucleosomes. These modifications can impact an RNA polymerase’s ability to find core promoters as well the activity of transcription factors. Transcriptional regulation involves cis-regulatory elements and trans-regulatory elements, which impact the function of the transcription initiation complex (TIC). Post-transcriptional regulation involves elements that impact the messenger RNA transcript itself. One of these mechanisms is microRNA (mi-R)-mediated mRNA decay, a process that leads to the degradation and subsequent downregulation of said transcript. Another mechanism that can impact the translated protein structure and/or function is alternative splicing.
2. General Aspects of the Molecular Mechanisms Underlying Adaptive Evolution in Vertebrates
Advances in sequencing, proteomics, and bioinformatic technologies enabled researchers to investigate the molecular mechanisms underlying adaptive variation. Sequence variation (e.g., indels (i.e., insertion–deletions) and point mutations) is one of these mechanisms (Figure 1). In some environmental contexts, sequence variation in key conserved sites can result in adaptions, but molecular implications of sequence variation/mutations vary depending on the type of conserved element impacted [5]. Mutations may arise via the insertion of transposable elements, which can result in variations in transcriptional regulation, DNA methylation patterns, and chromosome stability [6].
When mutations occur within the coding region, it has the potential to impact amino acid sequences near or within functional protein domains, and/or to affect post-transcriptional regulatory elements such as microRNAs (miRs). MiRs can be tissue-specific, can be developmental-time-specific, and may even be a response to stimuli [7][8][9]. MiR-mediated adaptive evolution can enable a lineage to gain or lose a regulatory sequence rather quickly, modifying regulatory patterns under novel pressure [8]. The modification of regulatory patterns leads to differential transcription. This can also occur due to (1) indirect factors (i.e., mutations leading to functional variation in proteins regulating transcription (e.g., transcription factors)) and (2) mutations in noncoding regulatory regions (e.g., enhancers and silencers) (Figure 1).
Another regulatory mechanism that can result in differential transcription is epigenetic modifications (Figure 1). Epigenetic changes are molecular modifications that affect genome structure, altering transcription rates by impacting the DNA binding ability of transcription factors and RNA polymerases [10]. Epigenetic modifications have been suggested to be essential for a rapid adaptive response to environmental factors [11]. Epigenetic changes do not always require novel mutations and are reversible (i.e., transient); thus, it can be an effective mechanism for differential gene regulation when organisms are subjected to sudden environmental pressures. Epigenetic variation can provide an immediate response to a sudden environmental change and can be transient.
Gene losses and losses of function can also give rise to evolutionary novelties, which can be advantageous in some ecological contexts [12]. The idea that gene loss can result in an increase in fitness is a controversial topic. Non-functionality of a gene leads to loss of the protein or proteins coded by this gene, affecting associated phenotypes. Some regulatory elements (e.g., silencers and corepressors) that contribute to gene loss of function have been deemed evolutionary conserved units, implying that gene loss may be adaptive. Exploring the possibility of how gene loss can be advantageous by yielding alternative phenotypes could provide further insight into the complexities of adaptive evolution.
3. Adaptive Evolution in Response to Variation in Lighting Conditions
Visual systems are important sensory systems used for navigation and receiving environmental stimuli. Lighting conditions vary considerably across environments, thus successful colonization of new environments requires adaptation of visual systems. Ecological factors shape visual sensitivities and have an impact on the molecular mechanisms that drive the adaptive evolution of visual structures and/or regulatory mechanisms in vertebrates [5][13][14][15][16]. Adaptation to a novel lighting environment is a complex process that requires changes in lens transmittance and retinal cell structure (i.e., chromatophore ratios and opsin gene expression) [13][14][17][18]. Opsins are photoreceptor proteins that bind to photons, thus interacting directly with an environmental stimulus.
Adaptive variation affecting retinae structure in response to variation in lighting conditions has been observed across multiple vertebrate species. One of the most common mechanisms is modifications near or within the functional domains of photoreceptors [16][19][20]. A well-described adaptive phenomenon in this regard is spectral tuning. Spectral tuning occurs when sequence variations impact photoreceptor binding sites, modifying the wavelength of the absorbed light that can be detected by photoreceptors [5][21].
Mutations in rhodopsin genes (RH1 and RH2) have been observed to cause spectral shifts in fishes, reptiles, and marine mammals as an adaptive response to variation in aquatic lighting conditions or lighting variation associated with living on the forest floor [14][15][21][22][23][24][25]. Spectral tuning associated with mutations in opsin genes (LWS and OPN1LW) has been proposed as an adaptive mechanism in vision acuity in giraffes [26]. Spectral tuning in the opsins LWS and SWS2 has also been observed as an adaptive mechanism contributing to nocturnal or benthic vertebrates’ ability to navigate low-lighted environments [19][24][27][28]. Both in reptile and fish lineages, relaxed constraints, at times leading to pseudogenization or gene loss, have been observed across species living in dimly lit environments [20][21]. SW1 loss has been proposed to be a beneficial phenotype, contributing to improving opsin sensibility towards longer wavelengths [19]. The shifts in spectral sensitivity are postulated to have occurred due to an increase in crepuscular and/or nocturnal activities of these species. Conversely, spectral tuning in SWS1 was observed in nocturnal reptiles and was proposed as an adaptive mechanism for diurnality [21].
Other molecular mechanisms underlying adaptive variation in visual systems are regulatory changes. In various fish species, the differential expression of enzymes and photoreceptors (e.g., CYP27C1, SWS1, RH2, RHO, and SWS2), and mutations in regulatory regions are suggested to be important molecular mechanisms underlying adaptive visual variation [5][20][24][29][30][31][32][33]. In cichlids, mutations in non-coding regions, specifically miR and transcription binding sites, have been found to be associated with differential transcription of opsin genes [32]. This highlights how a better understanding of where regulatory elements bind in the genome can provide essential information on how differential transcription occurs.
Regulatory modifications resulting in variations in the photoreceptor expression patterns are also suggested as a molecular mechanism underlying visual adaptations to nocturnal/dimly lit environments. In cichlids from dimly lit environments, mutations impacting dimerization properties, subsequently affecting downstream pathways regulated by these receptors, have been observed and are proposed to be adaptive [20]. Regulatory changes have impacted the retinal structure of nocturnal vertebrates to exhibit a higher proportion of rod cells, typically expressing RH1 [15][32][34]. RH1 may be essential for visual adaptations to dimly lit environments [23]. An electric knifefish species from dimly lit environments has been shown to exhibit compensatory substitutions in RH1 thought to be involved in re-establishing the dimerization properties of RH1, compensating for a RH1 mutation associated with the visual defects they exhibit in humans [35].
Mutations in functional domains have also been observed in non-photoreceptor visual systems genes. In the forest-dwelling okapi LUM, a gene involved in the phototransduction process exhibits a mutation suggested to impact the protein’s interaction with collagen, subsequently affecting UV transmission in dimly lit environments [26]. Additionally, the functional enhancement of retinae cell surface proteins (GRK1 and SLC24A1) has been proposed as a part of the underlying mechanisms of adaptations of vision in nocturnal birds [36].
Adaptive variations in visual systems are complex and can also result in adaptions impacting other aspects of phototransduction processes. In teleost fishes (Teleostei), molecular variations resulting in metabolic changes have been suggested to be involved in these visual system adaptations. A mutation in ATPase VHA involved in the increase in acidification of fish blood cells, leading to an increase in oxygen production and subsequent secretion into the retinae, has been proposed as a key adaptation [37]. This could have contributed to the morphological adaptations within teleost retinas and the adaptive evolution of their visual systems.
As previously mentioned, nocturnal vertebrates exhibit a higher proportion of rod cells within their retinae. Conversely, relaxed constraints of the visual system loci have been suggested to be involved in the variation and degeneration of nocturnal vertebrates’ visual systems [14][38][39][40][41][42]. Specifically, photoreceptor genes such as RH1, RH2, SWS1, or SWS2 have been lost under relaxed constraints in some of these species [7][40][42]. However, a unique mechanism of transmutation has been suggested as a means for vertebrates to adapt back to diurnal environments [7][35]. In members of the reptilian Colubridae family, which lost retinae cone cells, re-gaining a function has been demonstrated to occur through transmutation, resulting in evolutionary modifications of rods to become cone-like in function [7].
Overall, the adaptive variation of vertebrate visual systems is underlined by molecular modifications in retinae. Adapting to varying lighting conditions is a complex process, where many regulatory and structural genes are involved. Research has focused mainly on functional variation within photoreceptors that directly interact with environmental stimuli. However, adaptive variation has also been observed in proteins involved in phototransduction processes and in the expression patterns of photoreceptors. Thus, future studies should also investigate the putative regulatory mechanism, which may impact the expression patterns of photoreceptors as well as the phototransduction process itself.
4. Adaptations for Colonization of Aquatic and Terrestrial Environments
Ancestral vertebrates inhabited oceans and, later, transitioned into terrestrial environments. Conversely, some vertebrates regressed toward aquatic environments from terrestrial environments. Multiple molecular mechanisms involved in the vertebrate transition from water to land have been identified [43][44]. Li et al. (2018) investigated the underlaying molecular mechanisms that allowed vertebrates to invade terrestrial habitats by comparing the walking catfish and non-air breathing catfish. Their findings suggested that genes involved in DNA repair, enzyme activation, and small GTPase regulator activity were part of the molecular mechanisms to overcome the increase in DNA damage in terrestrial environments and the variation in metabolic processes.
The colonization of terrestrial environments also required vertebrates to adapt to hypoxic conditions, terrestrial xenobiotics, and novel environmental stimuli. Gene expansions and tissue-specific regulatory modifications of myoglobin genes (MB), sulfotransferase genes (e.g., SULT6B1), and olfactory receptor genes (e.g., ORA1) have been suggested to be involved in these processes [44]. The tissue-specific differential transcription of genes associated with ion homeostasis, acid–base balance, hemoglobin genes, angiogenesis, elastic fiber formation genes, and mutations in the functional domains of MB are thought to have contributed to overcoming hypoxic conditions in terrestrial environments [44][45].
The transition of vertebrates back to aquatic environments presented a unique evolutionary challenge, as many traits advantageous in these environments had been lost. Interestingly, for some complex traits, the relaxed constraints of specific genes yielded phenotypes that are advantageous in the return to aquatic environments. Specifically, the miR-based downregulation of genes has been suggested to be involved in adaptive variation for diving and overcoming hypoxic conditions. Tissue-specific variation in the microRNome of the deep diving Weddell seals suggests an important role of post-transcriptional regulatory mechanisms in marine mammal adaption to the aquatic environment. The tissue-specific differential expression of miRs targeting genes associated with hypoxia tolerance, anti-apoptotic pathways, and nitric oxide signal transduction was observed in Weddell seals [9]. MiR-mediated post-transcriptional regulation has been shown to be involved in downregulation, a mechanism that results in a substantial decrease in the amount of protein translated, which mimics the gene being “turned down” or “off” (Figure 1A) [8]. This suggests that the functional loss of some genes could potentially yield advantageous traits in marine mammal diving adaptions [9].
Eukaryotic regulatory pathways are complex, and losing a protein in the pathway which modifies it could potentially yield an advantageous phenotype by modifying instead of losing the trait [46]. This is the case for matrix metalloproteinase (MMP12), epidermal and hair development genes (DSC1, DSG4, TGM5, GSDMA, LYG1, and LYG2), and keratin genes (KRT9 and KRT20). MMP12 is involved in extracellular matrix breakdown, and its loss impacts pulmonary elasticity in a way that allows marine mammals to renew ~90% of their air in a single breath, an advantageous trait for diving [46]. The loss of the aforementioned epidermal and hair developmental genes resulted in modifications of the respective developmental pathways, yielding alternative traits of a thicker epidermis and hair loss traits in cetaceans and sirenians, suggested to be advantageous for the aquatic environments [46][47][48].
Loss of other functional proteins has also been suggested to have yielded advantageous traits in marine vertebrates. Relaxed constraints and subsequent gene loss of some olfactory and taste receptors genes (e.g., GNAT3 and CALHM1) observed in marine birds and mammals have been suggested to be advantageous traits by masking the smell and taste imbued onto their prey by the seawater [49][50]. Another example is the relaxed constraint of erythrocyte specific enzyme (AMPD3), which causes a reduced affinity to oxygen and a threefold increase in ATP in erythrocytes, an advantageous trait for diving [46].
Mutations in proteins with regulatory functions in cellular pathways (ACAN, a growth inhibitor), osteogenesis, and gene regulation (PIT-1, HOXD11, HOXD12, HOXD13 and MLL) have been proposed to be molecular mechanisms involved in the adaptive variation of the cetacean body architecture. The lineage-specific mutations are predicted to impact the structure and functionality of these proteins [51]. As in other vertebrates, they result in a body size reduction as well as shortened limbs and trunk [52]. Mutations in the aforementioned transcriptional factors impact their regulatory functions (e.g., binding affinity to target genes, chromatin remodeling, and histone modification), consequently modifying the transcriptional patterns of their target genes [10][53]. Target genes include genes involved in determining vertebrate body size (e.g., ELK1, LHX3, and PITX1), suggesting that these mutations are involved in the adaptive variation of cetacean body size and skeletal morphology [10][52][54].
The functional variation and expansion of genes involved in multiple cellular functions have also been proposed as underlying molecular mechanisms involved in adaptions to aquatic environments. Mutations in cetacean ATPase (ATP8) have been suggested to result in metabolic adjustments beneficial in their marine environments [55]. Mutations in a reproductive gene (FSHR) are suggested to be involved in adaptations of marine mammals’ reproductive systems [47]. Finally, the expansions of genes involved in multiple cellular processes (i.e., cellular response, oxidative stress, oxidation reduction, and hydrogen peroxide response) have been identified in cetaceans, other aquatic mammals, and semi aquatic mammals. They are likely involved in adaptations to hypoxic conditions, aquatic pathogens, novel energetic metabolic demands and nervous system adaptations [47][56][57][58].
Transitions to terrestrial and aquatic environments are complex evolutionary processes that require variation associated with both environmental stimuli (e.g., xenobiotics) and navigating these novel environments (e.g., diving). Due to the complex regulatory dynamics of eukaryotic gene expression, modification or even loss of regulatory proteins have yielded phenotypes that are advantageous in specific environmental contexts. Thus, future research in this topic would benefit from approaches that investigate the role of specific biochemical pathways involved in relevant cellular functions and gene regulation.
This entry is adapted from the peer-reviewed paper 10.3390/genes14020416
References
- Beldade, P.; Mateus, A.R.; Keller, R.A. Evolution and molecular mechanisms of adaptive developmental plasticity. Mol. Ecol. 2011, 20, 1347–1363.
- Olson-Manning, C.F.; Wagner, M.R.; Mitchell-Olds, T. Adaptive evolution: Evaluating empirical support for theoretical predictions. Nat. Rev. Genet. 2012, 13, 867–877.
- Wang, B.; Chen, L.; Wang, W. Genomic insights into ruminant evolution: From past to future prospects. Zool. Res. 2019, 40, 476–487.
- Slodkowicz, G.; Goldman, N. Integrated structural and evolutionary analysis reveals common mechanisms underlying adaptive evolution in mammals. Proc. Natl. Acad. Sci. USA 2020, 117, 5977–5986.
- Carleton, K.L.; Escobar-Camacho, D.; Stieb, S.M.; Cortesi, F.; Marshall, N.J. Seeing the rainbow: Mechanisms underlying spectral sensitivity in teleost fishes. J. Exp. Biol. 2020, 223, jeb193334.
- Da Silva, F.A.; Corrêa Guimarães, E.M.; Carvalho, N.; Ferreira, A.; Schneider, C.H.; Carvalho-Zilse, G.A.; Feldberg, E.; Gross, M. Transposable DNA Elements in Amazonian Fish: From Genome Enlargement to Genetic Adaptation to Stressful Environments. Cytogenet. Genome Res. 2020, 160, 148–155.
- Perry, B.W.; Card, D.C.; McGlothlin, J.W.; Pasquesi, G.I.M.; Adams, R.H.; Schield, D.R.; Hales, N.R.; Corbin, A.B.; Demuth, J.P.; Hoffmann, F.G.; et al. Molecular Adaptations for Sensing and Securing Prey and Insight into Amniote Genome Diversity from the Garter Snake Genome. Genome Biol. Evol. 2018, 10, 2110–2129.
- Luo, J.; Wang, Y.; Yuan, J.; Zhao, Z.; Lu, J. MicroRNA duplication accelerates the recruitment of new targets during vertebrate evolution. RNA 2018, 24, 787–802.
- Penso-Dolfin, L.; Haerty, W.; Hindle, A.; Di Palma, F. microRNA profiling in the Weddell seal suggests novel regulatory mechanisms contributing to diving adaptation. BMC Genom. 2020, 21, 303.
- Li, K.; Sun, X.; Chen, M.; Sun, Y.; Tian, R.; Wang, Z. Evolutionary changes of Hox genes and relevant regulatory factors provide novel insights into mammalian morphological modifications. Integr. Zool. 2018, 13, 21–35.
- Sun, Y.B.; Zhang, Y.; Wang, K. Perspectives on studying molecular adaptations of amphibians in the genomic era. Zool. Res. 2020, 41, 351–364.
- Lin, Q.; Fan, S.; Zhang, Y.; Xu, M.; Zhang, H.; Yang, Y.; Xia, W.; Liu, C.; Zhu, W.; Wang, H.; et al. The seahorse genome and the evolution of its specialized morphology. Nature 2016, 540, 395–399.
- Bloch, N.I. The evolution of opsins and color vision: Connecting genotype to a complex phenotype. Acta Biol. Colomb. 2016, 21, 481–494.
- Bian, C.; Huang, Y.; Li, J.; You, X.; Yi, Y.; Ge, W.; Shi, Q. Divergence, evolution and adaptation in ray-finned fish genomes. Sci. China Life Sci. 2019, 62, 1003–1018.
- Katti, C.; Stacey-Solis, M.; Coronel-Rojas, N.A.; Davies, W.I.L. The Diversity and Adaptive Evolution of Visual Photopigments in Reptiles. Front. Ecol. Evol. 2019, 7, 352.
- Musilova, Z.; Salzburger, W.; Cortesi, F. The Visual Opsin Gene Repertoires of Teleost Fishes: Evolution, Ecology, and Function. Annu. Rev. Cell Dev. Biol. 2021, 37, 441–468.
- Torres-Dowdall, J.; Pierotti, M.E.R.; Härer, A.; Karagic, N.; Woltering, J.M.; Henning, F.; Elmer, K.R.; Meter, A. Rapid and Parallel Adaptive Evolution of the Visual System of Neotropical Midas Cichlid Fishes. Mol. Biol. Evol. 2017, 34, 2469–2485.
- Lin, J.J.; Wang, F.Y.; Li, W.H.; Wang, T.Y. The rises and falls of opsin genes in 59 ray-finned fish genomes and their implications for environmental adaptation. Sci. Rep. 2017, 7, 15568.
- Carleton, K.L.; Dalton, B.E.; Escobar-Camacho, D.; Nandamuri, S.P. Proximate and ultimate causes of variable visual sensitivities: Insights from cichlid fish radiations. Genesis 2016, 54, 299–325.
- Hauser, F.E.; Ilves, K.L.; Schott, R.K.; Alvi, E.; López-Fernández, H.; Chang, B.S.W. Evolution, inactivation and loss of short wavelength-sensitive opsin genes during the diversification of Neotropical cichlids. Mol. Ecol. 2021, 30, 1688–1703.
- Cai, Y.; Fan, Y.; Yue, Y.; Li, P.; Yan, J.; Zhou, K. Molecular Evolution of Visual Opsin Genes during the Behavioral Shifts between Different Photic Environments in Geckos. Asian Herpetol. Res. 2021, 12, 280–288.
- Dungan, S.Z.; Kosyakov, A.; Chang, B.S. Spectral Tuning of Killer Whale (Orcinus orca) Rhodopsin: Evidence for Positive Selection and Functional Adaptation in a Cetacean Visual Pigment. Mol. Biol. Evol. 2016, 33, 323–336.
- Zheng, S.; Shao, F.; Tao, W.; Liu, Z.; Long, J.; Wang, X.; Zhang, S.; Zhao, Q.; Carleton, K.L.; Kocher, T.D. Chromosome-level assembly of southern catfish (Silurus meridionalis) provides insights into visual adaptation to nocturnal and benthic lifestyles. Mol. Ecol. Resour. 2021, 21, 1575–1592.
- Zhang, Z.; Liu, Y.; Zhang, W.; Du, X.; Liu, J. Benthic visual adaptation by fine-tuning light sensitivity in Japanese flounder (Paralichthys olivaceus). Front. Mar. Sci. 2022, 9, 1019660.
- Ito, R.K.; Harada, S.; Tabata, R.; Watanabe, K. Molecular evolution and convergence of the rhodopsin gene in Gymnogobius, a goby group having diverged into coastal to freshwater habitats. J. Evol. Biol. 2021, 35, 333–346.
- Ishengoma, E.; Agaba, M.; Cavener, D.R. Evolutionary analysis of vision genes identifies potential drivers of visual differences between giraffe and okapi. PeerJ 2017, 5, e3145.
- Wu, Y.; Hadly, E.A.; Teng, W.; Hao, Y.; Liang, W.; Liu, Y.; Wang, H. Retinal transcriptome sequencing sheds light on the adaptation to nocturnal and diurnal lifestyles in raptors. Sci. Rep. 2016, 6, 33578.
- Wu, Y.; Wang, H.; Hadly, E.A. Invasion of Ancestral Mammals into Dim-light Environments Inferred from Adaptive Evolution of the Phototransduction Genes. Sci. Rep. 2017, 7, 46542.
- Luehrmann, M.; Stieb, S.M.; Carleton, K.L.; Pietzker, A.; Cheney, K.L.; Marshall, N.J. Short-term colour vision plasticity on the reef: Changes in opsin expression under varying light conditions differ between ecologically distinct fish species. J. Exp. Biol. 2018, 221, jeb175281.
- Rennison, D.J.; Owens, G.L.; Heckman, N.; Schluter, D.; Veen, T. Rapid adaptive evolution of colour vision in the threespine stickleback radiation. Proc. Biol. Sci. 2016, 283, 20160242.
- Mehta, T.K.; Koch, C.; Nash, W.; Knaack, S.A.; Sudhakar, P.; Olbei, M.; Bastkowski, S.; Penso-Dolfin, L.; Korcsmaros, T.; Haerty, W.; et al. Evolution of regulatory networks associated with traits under selection in cichlids. Genome Biol. 2021, 22, 25.
- Mehta, T.K.; Penso-Dolfin, L.; Nash, W.; Roy, S.; Di-Palma, F.; Haerty, W. Evolution of miRNA-Binding Sites and Regulatory Networks in Cichlids. Mol. Biol. Evol. 2022, 39, 146.
- Wilwert, E.; Etienne, R.S.; van de Zande, L.; Maan, M.E. Contribution of opsins and chromophores to cone pigment variation across populations of Lake Victoria cichlids. J. Fish Biol. 2022, 101, 365–377.
- Mohun, S.M.; Davies, W.I.L. The Evolution of Amphibian Photoreception. Front. Ecol. Evol. 2019, 7, 321.
- van Nynatten, A.; Janzen, F.H.; Brochu, K.; Maldonado-Ocampo, J.A.; Crampton, W.G.R.; Chang, B.S.W. To see or not to see: Molecular evolution of the rhodopsin visual pigment in neotropical electric fishes. Proc. Biol. Sci. 2019, 286, 20191182.
- Wu, Y. Widespread nocturnality of living birds stemming from their common ancestor. BMC Evol. Biol. 2019, 19, 189.
- Damsgaard, C.; Lauridsen, H.; Harter, T.S.; Kwan, G.T.; Thomsen, J.S.; Funder, A.M.; Supuran, C.T.; Tresguerres, M.; Matthews, P.G.; Brauner, C.J. A novel acidification mechanism for greatly enhanced oxygen supply to the fish retina. eLife 2020, 9, e58995.
- Partha, R.; Chauhan, B.K.; Ferreira, Z.; Robinson, J.D.; Lathrop, K.; Nischal, K.K.; Chikina, M.; Ckarmk, N.L. Subterranean mammals show convergent regression in ocular genes and enhancers, along with adaptation to tunneling. eLife 2017, 6, e25884.
- Borges, R.; Fonseca, J.; Gomes, C.; Johnson, W.E.; O’Brien, S.J.; Zhang, G.; Gilbers, M.T.P.; Jarvis, E.D.; Atunes, A. Avian Binocularity and Adaptation to Nocturnal Environments: Genomic Insights from a Highly Derived Visual Phenotype. Genome Biol. Evol. 2019, 11, 2244–2255.
- Baldwin, M.W.; Ko, M.C. Functional evolution of vertebrate sensory receptors. Horm. Behav. 2020, 124, 104771.
- Jiang, M.; Shi, L.; Li, X.; Dong, Q.; Sun, H.; Du, Y.; Zhang, Y.; Shao, T.; Cheng, H.; Chen, W.; et al. Genome-wide adaptive evolution to underground stresses in subterranean mammals: Hypoxia adaption, immunity promotion, and sensory specialization. Ecol. Evol. 2020, 10, 7377–7388.
- Zhao, Y.; Chen, H.; Li, C.; Chen, S.; Xiao, H. Comparative Transcriptomics Reveals the Molecular Genetic Basis of Cave Adaptability in Sinocyclocheilus Fish Species. Front. Ecol. Evol. 2020, 8, 636503.
- Yuan, Z.; Liu, S.; Zhou, T.; Tian, C.; Bao, L.; Dunham, R.; Liu, Z. Comparative genome analysis of 52 fish species suggests differential associations of repetitive elements with their living aquatic environments. BMC Genom. 2018, 19, 141.
- Li, N.; Bao, L.; Zhou, T.; Yuan, Z.; Liu, S.; Dunham, R.; Li, Y.; Wang, K.; Xu, X.; Jin, Y.; et al. Genome sequence of walking catfish (Clarias batrachus) provides insights into terrestrial adaptation. BMC Genom. 2018, 19, 952.
- Tian, R.; Yin, D.; Liu, Y.; Seim, I.; Xu, S.; Yang, G. Adaptive Evolution of Energy Metabolism-Related Genes in Hypoxia-Tolerant Mammals. Front. Genet. 2017, 8, 205.
- Sharma, V.; Hecker, N.; Roscito, J.G.; Foerster, L.; Langer, B.E.; Hiller, M. A genomics approach reveals insights into the importance of gene losses for mammalian adaptations. Nat. Commun. 2018, 9, 1215.
- Zhou, X.; Sun, D.; Guang, X.; Ma, S.; Fang, X.; Mariotti, M.; Nielsen, R.; Gladyshev, V.N.; Yang, G. Molecular Footprints of Aquatic Adaptation Including Bone Mass Changes in Cetaceans. Genome Biol. Evol. 2018, 10, 967–975.
- Zhang, X.; Chi, H.; Li, G.; Irwin, D.M.; Zhang, S.; Rossiter, S.J.; Liu, Y. Parallel Independent Losses of G-Type Lysozyme Genes in Hairless Aquatic Mammals. Genome Biol. Evol. 2021, 13, evab201.
- Chikina, M.; Robinson, J.D.; Clark, N.L. Hundreds of Genes Experienced Convergent Shifts in Selective Pressure in Marine Mammals. Mol. Biol. Evol. 2016, 33, 2182–2192.
- Silva, M.C.; Chibucos, M.; Munro, J.B.; Daugherty, S.; Coelho, M.M.; Silva, J.C. Signature of adaptive evolution in olfactory receptor genes in Cory’s Shearwater supports molecular basis for smell in procellariiform seabirds. Sci. Rep. 2020, 10, 543.
- Sun, D.; Zhou, X.; Yu, Z.; Xu, S.; Seim, I.; Yang, G. Accelerated evolution and diversifying selection drove the adaptation of cetacean bone microstructure. BMC Evol. Biol. 2019, 19, 194.
- Sun, Y.; Liu, Y.; Sun, X.; Lin, Y.; Yin, D.; Xu, S.; Yang, G. Insights into body size variation in cetaceans from the evolution of body-size-related genes. BMC Evol. Biol. 2019, 19, 157.
- Li, J.; Shang, S.; Fang, N.; Zhu, Y.; Zhang, J.; Irwin, D.M.; Zhang, S.; Wang, Z. Accelerated Evolution of Limb-Related Gene Hoxd11 in the Common Ancestor of Cetaceans and Ruminants (Cetruminantia). G3 (Bethesda Md.) 2020, 10, 515–524.
- Li, G.; Wei, H.; Bi, J.; Ding, X.; Li, L.; Xu, S.; Yang, G.; Ren, W. Insights into Dietary Switch in Cetaceans: Evidence from Molecular Evolution of Proteinases and Lipases. J. Mol. Evol. 2020, 88, 521–535.
- Mori, S.; Matsunami, M. Signature of positive selection in mitochondrial DNA in Cetartiodactyla. Genes Genet. Syst. 2018, 93, 65–73.
- Tian, R.; Geng, Y.; Yang, Y.; Seim, I.; Yang, G. Oxidative stress drives divergent evolution of the glutathione peroxidase (GPX) gene family in mammals. Integr. Zool. 2021, 16, 696–711.
- Wang, L.; Zhou, S.; Lyu, T.; Shi, L.; Dong, Y.; He, S.; Zhang, H. Comparative Genome Analysis Reveals the Genomic Basis of Semi-Aquatic Adaptation in American Mink (Neovison vison). Animals 2022, 12, 2385.
- Qu, Y.; Chen, C.; Chen, X.; Hao, Y.; She, H.; Wang, M.; Ericson, P.; Lin, H.; Cai, T.; Song, G.; et al. The evolution of ancestral and species-specific adaptations in snowfinches at the Qinghai-Tibet Plateau. Proc. Natl. Acad. Sci. USA 2021, 118, e2012398118.
This entry is offline, you can click here to edit this entry!
