Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
In higher eukaryotes, the regulation of developmental gene expression is determined by enhancers, which are often located at a large distance from the promoters they regulate. Therefore, the architecture of chromosomes and the mechanisms that determine the functional interaction between enhancers and promoters are of decisive importance in the development of organisms. Mammals and the model animal Drosophila have homologous key architectural proteins and similar mechanisms in the organization of chromosome architecture.
- architectural C2H2 proteins
- CTCF
- Pita
- Su(Hw)
- boundary
1. Introduction
Differential expression of developmental genes in higher eukaryotes has led to a significant complication of the regulatory systems that control gene expression. Several promoters and dozens of enhancers often control expression of a single gene, and enhancers in some cases are hundreds of thousands of base pairs away from their target promoters [1][2]. The understanding of chromosome architecture and interactions between enhancers and promoters in higher eukaryotes is changing significantly with the development of methods that allow higher-resolution identification of distant contacts in the genome [3][4]. New research methods make it possible to study in more detail the architecture of chromosomes in the nucleus and how long-distance interactions between regulatory elements form. With the appearance of CRISPR/Cas9 technology, approaches to genome editing have been greatly simplified, and every DNA sequence can thus be added or deleted in the regulatory region of interest in vivo [5].
At present, Drosophila is the most convenient model object for studying the mechanisms of the formation of chromosome architecture common to all higher eukaryotes. Genome editing techniques can effectively be used in Drosophila to easily change particular genes and regulatory sequences [6]. Thus, it is possible to study the functions of every gene and to create complex model systems in vivo. The small size of the Drosophila genome facilitates high-resolution genome-wide studies, which yield more accurate results.
2. Models of Distance Interactions between Regulatory Elements
Two models have recently been proposed to explain long-range interactions in the genome. The main model is based on the findings that originate from mammalian Hi-C and ChIP-seq studies and indicate that the cohesin complex, together with CTCF, forms most of the enhancer–promoter interactions and boundaries of topology-associated domains (TADs) [7][8][9][10]. Inactivation of the cohesin complex or CTCF results in partial disruption of chromosome organization in TADs [11][12][13]. The cohesin complex is highly conserved in eukaryotes, and its main function is to hold sister chromatids together during mitosis and meiosis [14][15]. The cohesin complex consists of four subunits, which form a ring around the two DNA strands by using the energy of ATP [15]. A cluster consisting of 11 zinc finger domains of the C2H2 type is a feature of the structure of the CTCF protein [16][17][18]. Five C2H2 domains of CTCF specifically bind to a 15 bp motif, which is conserved in animals and determines most of the functional properties of this architectural protein [19]. A conserved motif interacting with the cohesin complex was found at the N-terminus of human CTCF [20]. A classical model suggests that, once fixed on chromatin, the cohesin complex begins ATP-dependent DNA extrusion with the formation of a chromatin loop [21]. CTCF blocks the movement of the cohesin complex, thus leading to fixation of the boundaries of chromatin loops at the CTCF sites [22].
An alternative group of models is based on the studies of mammalian LIM domain-binding factor 1 (LDB1) [23], the Drosophila architectural C2H2 proteins [24][25], and the Drosophila proteins that preferentially regulate the activity of housekeeping promoters [26][27].
In mammals, the C-terminal domain of LDB1 interacts with DNA-binding transcription factors of the LIM family [23]. The N-terminal domain of LDB1 forms a stable homodimer [28] to maintain long-range interactions between enhancers and gene promoters [29][30].
In Drosophila, several architectural C2H2 proteins have been characterized and shown to preferentially bind to gene promoters and known insulators [17][24][25]. The architectural proteins of this group have clusters of C2H2 domains, some of which specifically bind to motifs of 12 to 18 bp in length [17][24][25]. Most of the Drosophila C2H2 architectural proteins have structured domains that form homodimers at the N-terminus [31][32][33]. Interestingly, unstructured homodimerization domains are found at the N-terminus in the CTCF proteins of various animals, including Drosophila and mammals [34]. The domain is required for functional activity of Drosophila CTCF (dCTCF) [35], while the role of similar domains in mammalian CTCFs remains unstudied. In Drosophila, dCTCF, Pita, and Su(Hw) are the best-characterized architectural C2H2 proteins and determine the activity of most of the known Drosophila insulators [36][37][38]. Binding sites for these proteins can support long-distance interactions between regulatory elements in model transgenic lines [33][39][40].
The CP190, Chromator, Z4, and BEAF proteins preferentially bind to insulators and promoters of housekeeping genes, which are at the boundaries of most Drosophila TADs [26][27][41][42][43]. The proteins interact with each other and contain homodimerization domains [44][45][46][47][48], suggesting their likely involvement in maintaining long-distance interactions. Like mammalian LDB1, CP190 is recruited to regulatory elements through interactions with DNA-binding transcription factors including dCTCF, Pita, and Su(Hw) [49].
Either model by itself cannot explain a number of experimental results. For example, it was shown using Micro-C that inactivation of CTCF or cohesin does not affect the formation of chromatin loops between regulatory elements in mouse embryonic stem cells [50]. On the other hand, alternative models do not explain how distant chromatin regions initially find each other to form a stable pairing, which is necessary for the organization of chromatin loops. The most obvious is a combination of the two models, which will explain most of the current experimental data in both mammals and Drosophila (Figure 1A). In Drosophila, ChIP-seq data show that motifs recognized by different architectural C2H2 proteins are combined in many insulators and promoters [51][52]. Recent studies in mammals showed that, in well-studied genomic regions, CTCF binds in cooperation with the other C2H2 proteins ZNF143, MAZ, and WIZ [53][54][55][56], which are involved in the formation of long-distance interactions. MAZ and WIZ were shown to interact with the cohesin complex [54][55]. The cohesin complex likely interacts with a large number of C2H2 proteins. It can be assumed that the movement of cohesin complexes is most efficiently blocked in the chromatin regions that are associated with groups of C2H2 proteins. As a result, cohesin brings the regulatory elements together in a space, and their pairing is additionally stabilized by multiple interactions between the homodimerized domains of C2H2 architectural proteins and their associated partner proteins, such as CP190, Z4, and Chromator.
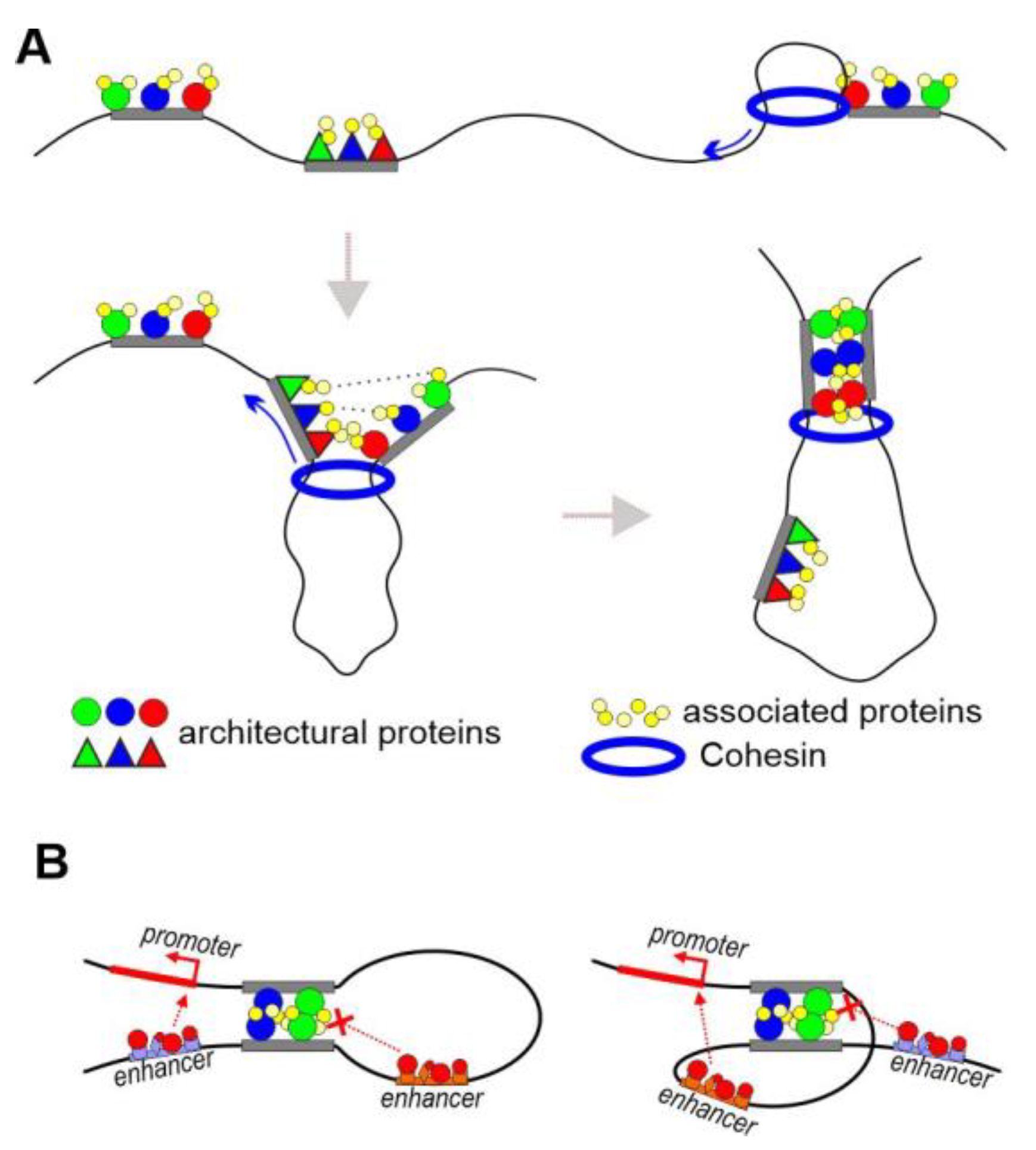
Figure 1. Combination of two models of distance interactions. (A) Local interaction between regulatory elements. Various combinations of architectural proteins bind to insulators or tethering elements. The same associated proteins (such as CP190, Z4, and Chromator) bind to different combinations of architectural proteins. The specificity of distance interactions between tethering elements/insulators is determined by the number of C2H2 proteins associated with different elements that are capable of interacting with each other. (B) Two copies of an insulator interact in head-to-head orientation.
The specificity and stability of the interaction between two regulatory elements is determined by the number of involved proteins whose domains are capable of forming homodimers (Figure 1A). Studies in transgenic Drosophila lines showed that two identical copies of any of the insulators tested pair in a head-to-head orientation [39][57]. When two identical insulators were oriented head-to-head, the configuration of the resulting chromatin loop was favorable for the interaction between a promoter and an enhancer located outside the loop (Figure 1B). When the insulators were in the same orientation, the enhancer could only stimulate the promoter when it was inside the loop. Such orientation-dependent interaction between identical copies of insulators is consistent with the model that regulatory elements consist of binding sites for several C2H2 architectural proteins, each of which can support long-distance interactions via its homodimerization domains. A direct consequence of the model is that inactivation of any architectural protein should not significantly affect the organization of chromosome architecture but may disrupt the individual local interactions between enhancers and promoters.
3. Current Models of Enhancer—Promoter Communication
Enhancers usually average about 500 bp in size and consist of combinations of motifs recognized by DNA-binding transcription factors (TFs), which suppress or activate enhancer activity (Figure 2A). [58]. Enhancers can be assembled into large modular super enhancers, which range in size from 5 to 50 kb [59]. The main function of enhancers is to mediate the recruitment of the mediator complex to promoters, resulting in transcriptional activation [60][61].
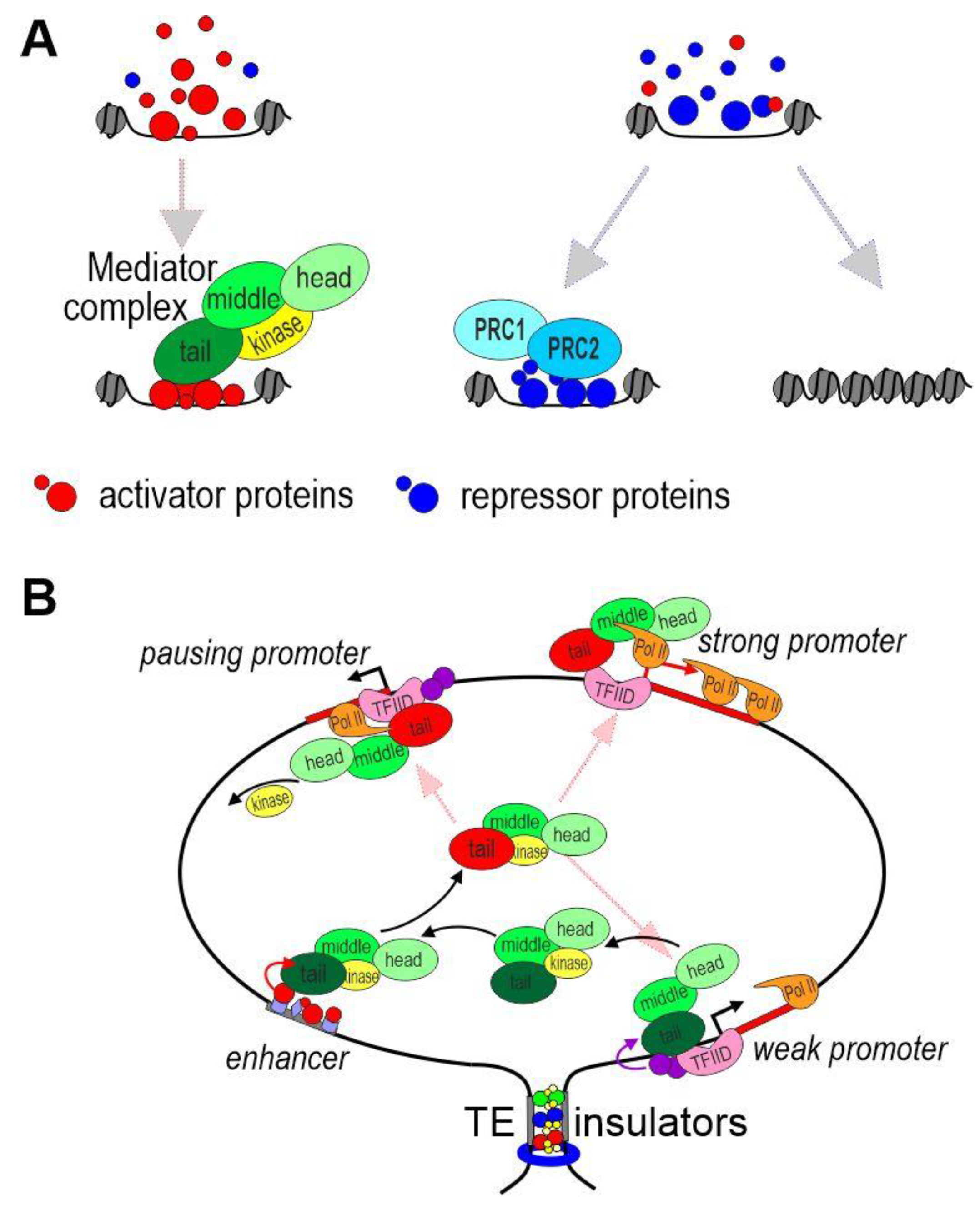
Figure 2. Model of promoter activation by an enhancer. (A) Activation or suppression of enhancers. The concentration of activators and repressors determines the fate of the enhancer in a particular nucleus. The mediator complex is recruited to the active enhancer. TFs can still bind to a repressed enhancer. In this case, Polycomb proteins play an important role in the suppression of enhancer activity. Alternatively, compaction of chromatin leads to dissociation of TFs from the enhancer. (B) Possible mechanism of functional interaction between an enhancer and promoters at a distance. Tethering elements or insulators form a chromatin loop that brings promoters into the active zone of the enhancer. The mediator complexes bind to the promoters located in the area of the enhancer. The level of transcription depends on the properties of a particular promoter.
The mediator complex is conserved in eukaryotes and consists of 26 subunits in mammals. The subunits are grouped in three modules, which are called the head, middle and tail (Figure 2A). A core part of the mediator interacts with the kinase module, which can function both as part of the complex and separately [60]. The head and middle modules provide interaction with RNA polymerase II; the tail module is responsible for the binding of the mediator with TFs on enhancers and the main TFIID complex on promoters [61] (Figure 2B). Binding to the mediator complex, the kinase module blocks its interaction with RNA polymerase II. The tail module is the most flexible and can take on various conformations [62][63]. The mediator complex binds to the non-phosphorylated carboxy-terminal domain (CTD) of RNA polymerase II, and the binding changes the conformation of the tail module. Next, RNA polymerase II is released from the complex with the mediator after CTD phosphorylation on the promoter to change the conformation of the tail module again. It is likely that different conformations of the tail module determine the specificity of binding of the mediator with TFs on enhancers or TFIID on promoters.
Several complexes with enzymatic activities are also recruited to enhancers: acetyltransferase (p300/CBP), methyltransferase (Mll3/Mll4/COMPASS), and deubiquitinase [64]. Mll3/Mll and p300/CBP are responsible for histone H3 monomethylation at lysine 4 (H3K4me1) and acetylation at lysine 27 (H3K27ac), respectively. The H3K27ac and H3K4me1 modifications of histone H3 are thought to reduce the stability of nucleosomes, resulting in the formation of open chromatin [65]. In addition, the enzymatic complexes can introduce modifications into TFs that bind to enhancers and gene promoters, thereby stimulating their activity [66][67]. For example, p300/CBP may play an important role in acetylation of the TFs involved in the pre-initiation complex formation [66]. Acetylation of different domains in the p53 protein usually positively regulates its activity [68]. Methylation of p53 at K327 increases its stability and ability to stimulate transcription [69]. There are other examples of the positive role of TF methylation and acetylation, but this area remains poorly studied in general.
In addition to transcription activators, repressors are recruited to enhancers to suppress their activity in cells where the enhancers should not function (Figure 2A). Complexes with deacetylase and, less commonly, demethylase activities are recruited to enhancers by repressors [70]. Deacetylation of TFs on enhancers probably decreases their ability to attract enzymatic and mediator complexes. In addition, histone deacetylation increases chromatin compaction, thereby reducing the ability of TFs to bind enhancers [71]. Thus, enhancer activity in a particular cell is determined by the concentration of TFs interacting with activator and repressor complexes (Figure 2A).
The Polycomb proteins play an important role in the suppression of enhancer activity [71][72][73]. Two main Polycomb complexes are known in Drosophila, of which one has ubiquitinating activity (Polycomb repression complex 1, PRC1) and the other has methyltransferase activity (Polycomb repression complex 2, PRC2) [73]. PRC1 and PRC2 can be recruited directly to enhancers and promoters through interactions with DNA-binding TFs [74]. A large number of variations in these two basic Polycomb complexes have been found in mammals, with them being determined by the need to finely regulate numerous groups of enhancers and promoters during development and cell differentiation [71]. The most studied mechanism of repression is the formation of inactive chromatin through the introduction of H3K27me3 and H2AK119ub modifications into nucleosomes mediated by Polycomb complexes [71][73]. Methylation and ubiquitination of key TFs is also a possible mechanism to suppress enhancers and promoters. For example, methylation of lysine 99 in the coactivator BRD4 negatively regulates its activity in transcription [75].
Recruitment of the Polycomb complexes to enhancers can lead to their transformation into silencers that repress transcription of adjacent genes [76][77][78]. Drosophila has well-characterized, specialized regulatory elements that specifically recruit PRC1 and PRC2 and they are called Polycomb response elements (PREs) [79]. Such regulatory elements can function as specific silencers, increasing the efficiency of the complete repression of the enhancers and promoters that should be completely turned off in a certain group of cells during development [80][81].
Two recent studies [82][83] investigated the compatibility of enhancers and promoters. It was found that enhancers preferentially activate weak promoters rather than strong promoters, which normally determine the transcription of housekeeping and cell cycle genes. In general, it was shown that most of the enhancers tested can activate almost every promoter. A lack of specificity of interactions between enhancers and promoters presumably increases the role of insulators and TADs in limiting enhancer–promoter interactions.
However, recent studies have shown that TADs do not block long-range interactions between enhancers and promoters [50]. It was shown using Drosophila transgenic model systems that chromatin loops formed by interacting insulators cannot effectively block the interaction between enhancers and promoters [40][84][85]. Thus, there are no strict structural restrictions to block the co-localization of enhancers and promoters belonging to different regulatory domains. Using micro-C, intense contacts were detected in the genome between certain genomic sites including enhancers, promoters, and insulators that do not coincide with TAD boundaries [50][86][87][88]. A special class of regulatory elements, called tethering elements, was isolated in Drosophila. The elements occur next to enhancers and promoters and form stable chromatin loops between them [86]. Ultra-high resolution microscopy showed that some functionally interacting enhancers and promoters are relatively far away from each other [3][89].
It can be assumed that mediator complexes are concentrated on enhancers as a result of multiple interactions between subunits of the tail module and unstructured domains of enhancer-associated TFs [61] (Figure 2B). In the next stage, the mediator leaves the enhancer as a result of a change in the conformation of the tail module. Conformational changes in the tail module are possibly a result of methylation (Mll3/Mll4/COMPASS?), acetylation (p300/CBP?), or phosphorylation (the kinase module?) of subunits of the mediator complex. However, this issue has not been studied as of yet. In the new conformation, the tail module has greater affinity for the TFIID complex on the promoter, resulting in pre-initiation complex formation and the recruitment of RNA polymerase II. The enhancer-bound p300/CBP complex can simultaneously acetylate TFs to activate them. Increasing concentrations of active forms of the mediator complex and TFs should stimulate the promoters located in a certain active zone around the enhancer [89][90]. It does not matter to such a trans-activation mechanism whether the enhancer and promoter are in close contact, interact briefly, or are at some distance from each other. Interactions between insulators and/or tethering elements lead to the formation of chromatin loops, which form a region in which enhancers stimulate a specific group of promoters. In some cases, chromatin loops can reduce the likelihood of promoter localization in the nuclear region where the enhancer functions.
4. Interacting Insulators form an Autonomous Regulatory Domain of the eve Gene
The regulation of the pair-rule gene even-skipped (eve) is one of the best studied in Drosophila (Figure 3A). [91][92][93][94]. Eve belongs to a group of primary pair-rule factors whose stripe-pattern expression starts in early embryonic development [95][96]. The eve gene is in the center of a 16 kb domain surrounded by housekeeping genes, which are active in all cells.
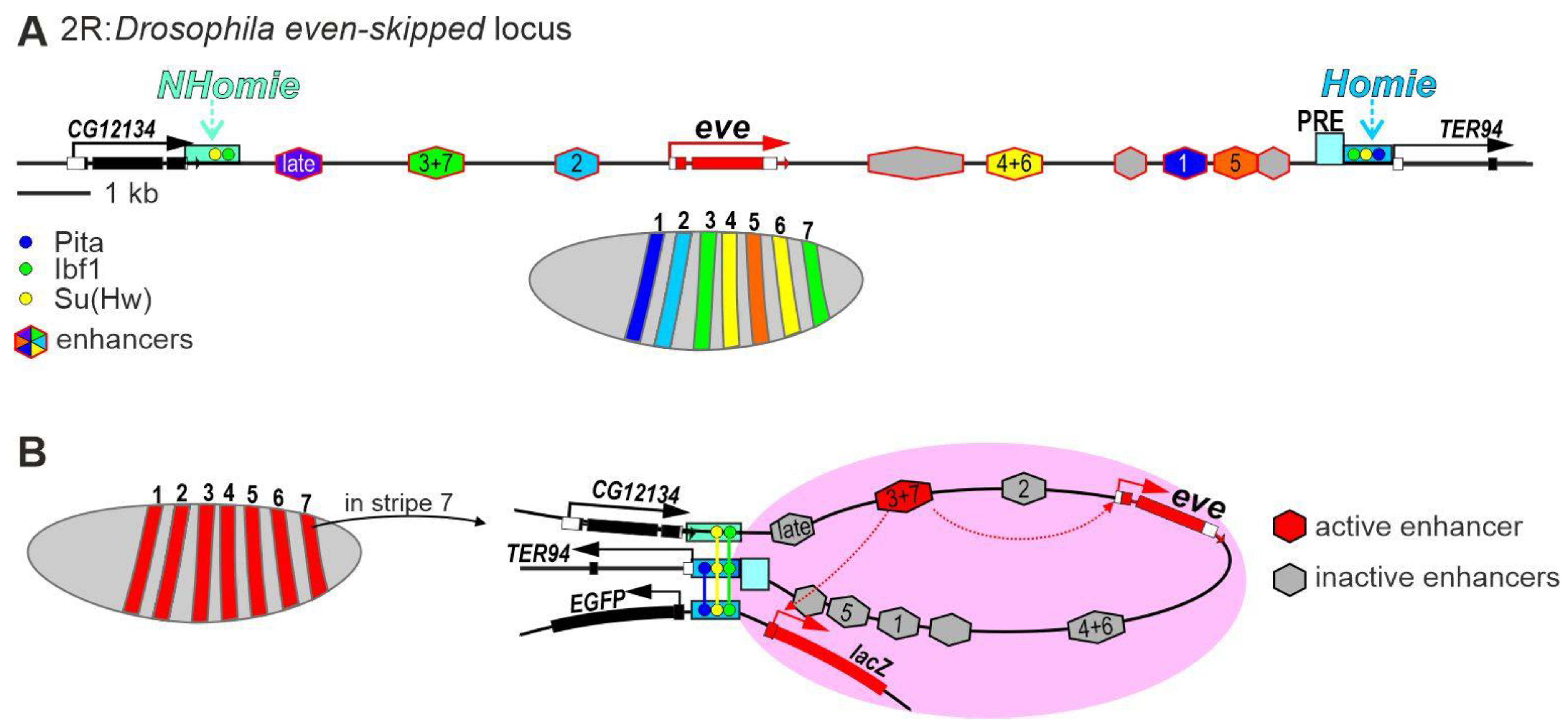
Figure 3. Model of transcriptional regulation of the pair-rule gene eve in early Drosophila embryos. (A) Schematic representation of the eve regulatory region that is flanked by the Homie and NHomie insulators. (B) Transcriptional activation model of the endogenous eve gene and the reporter transgene in the stripe 7 of early embryos. The interaction between the Homie and NHomie insulators forms a zone in which the activated eve enhancer can stimulate transcription of the endogenous eve promoter and the reporter gene promoter. Identical copies of the Homie insulator located in the endogenous eve locus and the transgene interact in head-to-head orientation, which brings only the reporter located on the head side of the insulator into the active eve enhancer zone.
The body is divided into segments with certain morphological differences in Drosophila, like in all insects [97]. Segments formed at the embryonic stage are called parasegments (PSs). During the early development of an embryo, 14 PSs are formed, corresponding to anatomical structures of the larva. PSs are initially determined by the products of the maternal genes Bicoid (Bcd), Hunchback (Hb), and Caudal (Cad), which precisely regulate the expression levels of gap group genes, including hunchback (hb), Kruppel (Kr), knisps (kni), and giant (gt) [98][99][100][101]. In early embryos, the maternal and gap genes cooperatively regulate the expression of a large group of pair-rule genes, including eve and fushi tarazu (ftz) [102][103]. The eve gene is expressed in seven broad stripes along the anteroposterior (AP) axis of the embryo during its early development (Figure 3A). At this stage, eve expression is controlled by five enhancers that are active in separate stripes [95][96]. The stripes that express eve subsequently become thinner with clear anterior and posterior borders [104]. Expression of the eve gene at this stage is controlled by a single enhancer, which is bound with the early pair-rule proteins paired, runt, and sloppy-paired [105]. At late stages of embryonic development, eve expression loses its characteristic pattern and is controlled by several tissue-specific enhancers.
The eve enhancers contain binding sites for ubiquitous transcriptional activators, such as STAT and Zelda (Zld), and the maternal Bicoid activator [70][106][107][108]. Repression of the enhancers is controlled by the Kr, Kni, and Gt proteins, which recruit the CtBP repressor complex [70]. CtBP-dependent repressor complexes have deacetylase activity. Finally, the Hb protein can recruit activators or repressors to the enhancers, depending on the nearby partner proteins [109]. For example, the stripe 3 + 7 enhancer is stimulated by the activators Zld and STAT and repressed by Hb and Kni [107][108]. At the same time, the stripe 2 enhancer is controlled positively by Zld, Hb, and Bcd and negatively by Gt and Kr.
Each stripe enhancer has a specific set of activator and repressor motifs, which are arranged in a specific sequence and orientation. Each stripe enhancer shows more efficient recruitment of activator (acetylase activity) or suppressor (deacetylase activity) complexes, depending on the concentration of gap repressors in the nucleus. TF acetylation/deacetylation is likely to stabilize the active/inactive status of each stripe enhancer. Deacetylation of nucleosomes also leads to the formation of more stable local chromatin, which blocks the binding of activators to enhancers. This possibility is consistent with the finding that the Zelda and Hb proteins cannot stably bind to their sites on chromatin [110].
The complex regulatory region of the eve gene (Figure 3A) is flanked by housekeeping genes, which are expressed in all cells [111][112]. The housekeeping gene TER94 is on one side of the regulatory region of the eve gene and is actively transcribed in all cells. The other side is flanked by the 3′ region of the CG12134 gene, which shows ubiquitous but weaker expression.
A 368 bp insulator (Figure 3A) was found immediately upstream of the core promoter of the TER94 gene [111][112]. The insulator efficiently blocks the activity of embryonic enhancers in model transgenic lines. When the insulator was inserted into the P-transposon, the construct was found to preferentially integrate into the genomic region near the eve locus [111][112]. This effect is called homing and is explained as follows. When DNA of the P-transposon with the insulator is injected, proteins are assembled on the insulator to form a complex, which interacts with a similar complex on the endogenous insulator to increase the specific integration of the P-transposon. The insulator was therefore named Homie. The function of Homie in vivo is currently unknown since its deletion has not been obtained. It is likely that Homie performs many functions, one of which is to be the distal part of the TER94 gene promoter since deletion of the insulator significantly reduced TER94 expression in transgenic lines [111].
A PRE was found next to the insulator (Figure 3A); its function is to negatively regulate the eve gene enhancers at the late stages of embryogenesis [111]. Homie was assumed to protect TER94 expression from the PRE, which represses TER94 transcription in oocytes and late embryos in transgenic lines [111]. A second insulator (Figure 3A), named new Homie (NHomie), was found between the 3′ UTR of the CG12134 gene and the regulatory region of the eve locus [113]. Interestingly, both insulators are bound with the Su(Hw) [114] and Ibf1/2 [115] proteins. The proteins can be involved in recruiting CP190 and Mod(mdg4)-67.2 to Homie and NHomie [115][116]. Homie additionally binds with Pita, which is another architectural C2H2 protein, and also interacts with CP190 [52][117]. Thus, the Homie insulator has binding sites for two architectural C2H2 proteins. In Micro-C studies, Homie and NHomie efficiently interacted to form a small TAD in embryos [86].
This entry is adapted from the peer-reviewed paper 10.3390/ijms24032855
References
- Andersson, R.; Sandelin, A. Determinants of Enhancer and Promoter Activities of Regulatory Elements. Nat. Rev. Genet. 2020, 21, 71–87.
- Furlong, E.E.M.; Levine, M. Developmental Enhancers and Chromosome Topology. Science 2018, 361, 1341–1345.
- Hafner, A.; Boettiger, A. The Spatial Organization of Transcriptional Control. Nat. Rev. Genet. 2022, 24, 53–68.
- Jerkovic, I.; Cavalli, G. Understanding 3D Genome Organization by Multidisciplinary Methods. Nat. Rev. Mol. Cell Biol. 2021, 22, 511–528.
- Wang, H.; La Russa, M.; Qi, L.S. CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem. 2016, 85, 227–264.
- Housden, B.E.; Perrimon, N. Cas9-Mediated Genome Engineering in Drosophila Melanogaster. Cold Spring Harb. Protoc. 2016, 2016, pdb-top086843.
- Sexton, T.; Yaffe, E.; Kenigsberg, E.; Bantignies, F.; Leblanc, B.; Hoichman, M.; Parrinello, H.; Tanay, A.; Cavalli, G. Three-Dimensional Folding and Functional Organization Principles of the Drosophila Genome. Cell 2012, 148, 458–472.
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological Domains in Mammalian Genomes Identified by Analysis of Chromatin Interactions. Nature 2012, 485, 376–380.
- Rao, S.S.P.; Huntley, M.H.; Durand, N.C.; Stamenova, E.K.; Bochkov, I.D.; Robinson, J.T.; Sanborn, A.L.; Machol, I.; Omer, A.D.; Lander, E.S.; et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell 2014, 159, 1665–1680.
- Ulianov, S.V.; Khrameeva, E.E.; Gavrilov, A.A.; Flyamer, I.M.; Kos, P.; Mikhaleva, E.A.; Penin, A.A.; Logacheva, M.D.; Imakaev, M.V.; Chertovich, A.; et al. Active Chromatin and Transcription Play a Key Role in Chromosome Partitioning into Topologically Associating Domains. Genome Res. 2016, 26, 70–84.
- Nora, E.P.; Goloborodko, A.; Valton, A.-L.; Gibcus, J.H.; Uebersohn, A.; Abdennur, N.; Dekker, J.; Mirny, L.A.; Bruneau, B.G. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 2017, 169, 930–944.e22.
- Rao, S.S.P.; Huang, S.-C.; Glenn St Hilaire, B.; Engreitz, J.M.; Perez, E.M.; Kieffer-Kwon, K.-R.; Sanborn, A.L.; Johnstone, S.E.; Bascom, G.D.; Bochkov, I.D.; et al. Cohesin Loss Eliminates All Loop Domains. Cell 2017, 171, 305–320.e24.
- Schwarzer, W.; Abdennur, N.; Goloborodko, A.; Pekowska, A.; Fudenberg, G.; Loe-Mie, Y.; Fonseca, N.A.; Huber, W.; Haering, C.H.; Mirny, L.; et al. Two Independent Modes of Chromatin Organization Revealed by Cohesin Removal. Nature 2017, 551, 51–56.
- Dorsett, D. The Many Roles of Cohesin in Drosophila Gene Transcription. Trends Genet. 2019, 35, 542–551.
- Davidson, I.F.; Peters, J.-M. Genome Folding through Loop Extrusion by SMC Complexes. Nat. Rev. Mol. Cell Biol. 2021, 22, 445–464.
- Arzate-Mejía, R.G.; Recillas-Targa, F.; Corces, V.G. Developing in 3D: The Role of CTCF in Cell Differentiation. Development 2018, 145, dev137729.
- Maksimenko, O.G.; Fursenko, D.V.; Belova, E.V.; Georgiev, P.G. CTCF As an Example of DNA-Binding Transcription Factors Containing Clusters of C2H2-Type Zinc Fingers. Acta Nat. 2021, 13, 31–46.
- Ohlsson, R.; Renkawitz, R.; Lobanenkov, V. CTCF Is a Uniquely Versatile Transcription Regulator Linked to Epigenetics and Disease. Trends Genet. 2001, 17, 520–527.
- Hashimoto, H.; Wang, D.; Horton, J.R.; Zhang, X.; Corces, V.G.; Cheng, X. Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA. Mol. Cell 2017, 66, 711–720.e3.
- Li, Y.; Haarhuis, J.H.I.; Sedeño Cacciatore, Á.; Oldenkamp, R.; van Ruiten, M.S.; Willems, L.; Teunissen, H.; Muir, K.W.; de Wit, E.; Rowland, B.D.; et al. The Structural Basis for Cohesin-CTCF-Anchored Loops. Nature 2020, 578, 472–476.
- Oldenkamp, R.; Rowland, B.D. A Walk through the SMC Cycle: From Catching DNAs to Shaping the Genome. Mol. Cell 2022, 82, 1616–1630.
- Fudenberg, G.; Imakaev, M.; Lu, C.; Goloborodko, A.; Abdennur, N.; Mirny, L.A. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016, 15, 2038–2049.
- Krivega, I.; Dean, A. Chromatin Looping as a Target for Altering Erythroid Gene Expression. Ann. N. Y. Acad. Sci. 2016, 1368, 31–39.
- Kyrchanova, O.; Georgiev, P. Mechanisms of Enhancer-Promoter Interactions in Higher Eukaryotes. Int. J. Mol. Sci. 2021, 22, 671.
- Maksimenko, O.; Kyrchanova, O.; Klimenko, N.; Zolotarev, N.; Elizarova, A.; Bonchuk, A.; Georgiev, P. Small Drosophila Zinc Finger C2H2 Protein with an N-Terminal Zinc Finger-Associated Domain Demonstrates the Architecture Functions. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194446.
- Ramírez, F.; Bhardwaj, V.; Arrigoni, L.; Lam, K.C.; Grüning, B.A.; Villaveces, J.; Habermann, B.; Akhtar, A.; Manke, T. High-Resolution TADs Reveal DNA Sequences Underlying Genome Organization in Flies. Nat. Commun. 2018, 9, 189.
- Wang, Q.; Sun, Q.; Czajkowsky, D.M.; Shao, Z. Sub-Kb Hi-C in D. Melanogaster Reveals Conserved Characteristics of TADs between Insect and Mammalian Cells. Nat. Commun. 2018, 9, 188.
- Wang, H.; Kim, J.; Wang, Z.; Yan, X.-X.; Dean, A.; Xu, W. Crystal Structure of Human LDB1 in Complex with SSBP2. Proc. Natl. Acad. Sci. USA 2020, 117, 1042–1048.
- Krivega, I.; Dale, R.K.; Dean, A. Role of LDB1 in the Transition from Chromatin Looping to Transcription Activation. Genes Dev. 2014, 28, 1278–1290.
- Deng, W.; Lee, J.; Wang, H.; Miller, J.; Reik, A.; Gregory, P.D.; Dean, A.; Blobel, G.A. Controlling Long-Range Genomic Interactions at a Native Locus by Targeted Tethering of a Looping Factor. Cell 2012, 149, 1233–1244.
- Bonchuk, A.N.; Boyko, K.M.; Nikolaeva, A.Y.; Burtseva, A.D.; Popov, V.O.; Georgiev, P.G. Structural Insights into Highly Similar Spatial Organization of Zinc-Finger Associated Domains with a Very Low Sequence Similarity. Structure 2022, 30, 1004–1015.e4.
- Bonchuk, A.; Boyko, K.; Fedotova, A.; Nikolaeva, A.; Lushchekina, S.; Khrustaleva, A.; Popov, V.; Georgiev, P. Structural Basis of Diversity and Homodimerization Specificity of Zinc-Finger-Associated Domains in Drosophila. Nucleic Acids Res. 2021, 49, 2375–2389.
- Zolotarev, N.; Fedotova, A.; Kyrchanova, O.; Bonchuk, A.; Penin, A.A.; Lando, A.S.; Eliseeva, I.A.; Kulakovskiy, I.V.; Maksimenko, O.; Georgiev, P. Architectural Proteins Pita, Zw5,and ZIPIC Contain Homodimerization Domain and Support Specific Long-Range Interactions in Drosophila. Nucleic Acids Res. 2016, 44, 7228–7241.
- Bonchuk, A.; Kamalyan, S.; Mariasina, S.; Boyko, K.; Popov, V.; Maksimenko, O.; Georgiev, P. N-Terminal Domain of the Architectural Protein CTCF Has Similar Structural Organization and Ability to Self-Association in Bilaterian Organisms. Sci. Rep. 2020, 10, 2677.
- Bonchuk, A.; Maksimenko, O.; Kyrchanova, O.; Ivlieva, T.; Mogila, V.; Deshpande, G.; Wolle, D.; Schedl, P.; Georgiev, P. Functional Role of Dimerization and CP190 Interacting Domains of CTCF Protein in Drosophila Melanogaster. BMC Biol. 2015, 13, 63.
- Melnikova, L.S.; Georgiev, P.G.; Golovnin, A.K. The Functions and Mechanisms of Action of Insulators in the Genomes of Higher Eukaryotes. Acta Nat. 2020, 12, 15–33.
- Matthews, N.E.; White, R. Chromatin Architecture in the Fly: Living without CTCF/Cohesin Loop Extrusion?: Alternating Chromatin States Provide a Basis for Domain Architecture in Drosophila. Bioessays 2019, 41, e1900048.
- Chen, D.; Lei, E.P. Function and Regulation of Chromatin Insulators in Dynamic Genome Organization. Curr. Opin. Cell Biol. 2019, 58, 61–68.
- Kyrchanova, O.; Chetverina, D.; Maksimenko, O.; Kullyev, A.; Georgiev, P. Orientation-Dependent Interaction between Drosophila Insulators Is a Property of This Class of Regulatory Elements. Nucleic Acids Res. 2008, 36, 7019–7028.
- Kyrchanova, O.; Maksimenko, O.; Stakhov, V.; Ivlieva, T.; Parshikov, A.; Studitsky, V.M.; Georgiev, P. Effective Blocking of the White Enhancer Requires Cooperation between Two Main Mechanisms Suggested for the Insulator Function. PLoS Genet. 2013, 9, e1003606.
- Cubeñas-Potts, C.; Rowley, M.J.; Lyu, X.; Li, G.; Lei, E.P.; Corces, V.G. Different Enhancer Classes in Drosophila Bind Distinct Architectural Proteins and Mediate Unique Chromatin Interactions and 3D Architecture. Nucleic Acids Res. 2017, 45, 1714–1730.
- Pal, K.; Forcato, M.; Jost, D.; Sexton, T.; Vaillant, C.; Salviato, E.; Mazza, E.M.C.; Lugli, E.; Cavalli, G.; Ferrari, F. Global Chromatin Conformation Differences in the Drosophila Dosage Compensated Chromosome X. Nat. Commun. 2019, 10, 5355.
- Dong, Y.; Avva, S.V.S.P.; Maharjan, M.; Jacobi, J.; Hart, C.M. Promoter-Proximal Chromatin Domain Insulator Protein BEAF Mediates Local and Long-Range Communication with a Transcription Factor and Directly Activates a Housekeeping Promoter in Drosophila. Genetics 2020, 215, 89–101.
- Bonchuk, A.; Denisov, S.; Georgiev, P.; Maksimenko, O. Drosophila BTB/POZ Domains of “Ttk Group” Can Form Multimers and Selectively Interact with Each Other. J. Mol. Biol. 2011, 412, 423–436.
- Gan, M.; Moebus, S.; Eggert, H.; Saumweber, H. The Chriz-Z4 Complex Recruits JIL-1 to Polytene Chromosomes, a Requirement for Interband-Specific Phosphorylation of H3S10. J. Biosci. 2011, 36, 425–438.
- Vogelmann, J.; Le Gall, A.; Dejardin, S.; Allemand, F.; Gamot, A.; Labesse, G.; Cuvier, O.; Nègre, N.; Cohen-Gonsaud, M.; Margeat, E.; et al. Chromatin Insulator Factors Involved in Long-Range DNA Interactions and Their Role in the Folding of the Drosophila Genome. PLoS Genet. 2014, 10, e1004544.
- Melnikova, L.S.; Kostyuchenko, M.V.; Georgiev, P.G.; Golovnin, A.K. The Chriz Protein Promotes the Recruitment of the Z4 Protein to the STAT-Dependent Promoters. Dokl. Biochem. Biophys. 2020, 490, 29–33.
- Melnikova, L.S.; Molodina, V.V.; Kostyuchenko, M.V.; Georgiev, P.G.; Golovnin, A.K. The BEAF-32 Protein Directly Interacts with Z4/Putzig and Chriz/Chromator Proteins in Drosophila Melanogaster. Dokl. Biochem. Biophys. 2021, 498, 184–189.
- Sabirov, M.; Popovich, A.; Boyko, K.; Nikolaeva, A.; Kyrchanova, O.; Maksimenko, O.; Popov, V.; Georgiev, P.; Bonchuk, A. Mechanisms of CP190 Interaction with Architectural Proteins in Drosophila Melanogaster. Int. J. Mol. Sci. 2021, 22, 12400.
- Hsieh, T.-H.S.; Cattoglio, C.; Slobodyanyuk, E.; Hansen, A.S.; Darzacq, X.; Tjian, R. Enhancer-Promoter Interactions and Transcription Are Largely Maintained upon Acute Loss of CTCF, Cohesin, WAPL or YY1. Nat. Genet. 2022, 54, 1919–1932.
- Kyrchanova, O.; Maksimenko, O.; Ibragimov, A.; Sokolov, V.; Postika, N.; Lukyanova, M.; Schedl, P.; Georgiev, P. The Insulator Functions of the Drosophila Polydactyl C2H2 Zinc Finger Protein CTCF: Necessity versus Sufficiency. Sci. Adv. 2020, 6, eaaz3152.
- Sabirov, M.; Kyrchanova, O.; Pokholkova, G.V.; Bonchuk, A.; Klimenko, N.; Belova, E.; Zhimulev, I.F.; Maksimenko, O.; Georgiev, P. Mechanism and Functional Role of the Interaction between CP190 and the Architectural Protein Pita in Drosophila Melanogaster. Epigenetics Chromatin 2021, 14, 16.
- Ortabozkoyun, H.; Huang, P.-Y.; Cho, H.; Narendra, V.; LeRoy, G.; Gonzalez-Buendia, E.; Skok, J.A.; Tsirigos, A.; Mazzoni, E.O.; Reinberg, D. CRISPR and Biochemical Screens Identify MAZ as a Cofactor in CTCF-Mediated Insulation at Hox Clusters. Nat. Genet. 2022, 54, 202–212.
- Xiao, T.; Li, X.; Felsenfeld, G. The Myc-Associated Zinc Finger Protein (MAZ) Works Together with CTCF to Control Cohesin Positioning and Genome Organization. Proc. Natl. Acad. Sci. USA 2021, 118, e2023127118.
- Justice, M.; Carico, Z.M.; Stefan, H.C.; Dowen, J.M. A WIZ/Cohesin/CTCF Complex Anchors DNA Loops to Define Gene Expression and Cell Identity. Cell Rep. 2020, 31, 107503.
- Zhou, Q.; Yu, M.; Tirado-Magallanes, R.; Li, B.; Kong, L.; Guo, M.; Tan, Z.H.; Lee, S.; Chai, L.; Numata, A.; et al. ZNF143 Mediates CTCF-Bound Promoter-Enhancer Loops Required for Murine Hematopoietic Stem and Progenitor Cell Function. Nat. Commun. 2021, 12, 43.
- Kyrchanova, O.; Georgiev, P. Chromatin Insulators and Long-Distance Interactions in Drosophila. FEBS Lett. 2014, 588, 8–14.
- Spitz, F.; Furlong, E.E.M. Transcription Factors: From Enhancer Binding to Developmental Control. Nat. Rev. Genet. 2012, 13, 613–626.
- Blobel, G.A.; Higgs, D.R.; Mitchell, J.A.; Notani, D.; Young, R.A. Testing the Super-Enhancer Concept. Nat. Rev. Genet. 2021, 22, 749–755.
- Luyties, O.; Taatjes, D.J. The Mediator Kinase Module: An Interface between Cell Signaling and Transcription. Trends Biochem. Sci. 2022, 47, 314–327.
- Richter, W.F.; Nayak, S.; Iwasa, J.; Taatjes, D.J. The Mediator Complex as a Master Regulator of Transcription by RNA Polymerase II. Nat. Rev. Mol. Cell Biol. 2022, 23, 732–749.
- Chen, D.; McManus, C.E.; Radmanesh, B.; Matzat, L.H.; Lei, E.P. Temporal Inhibition of Chromatin Looping and Enhancer Accessibility during Neuronal Remodeling. Nat. Commun. 2021, 12, 6366.
- Robinson, P.J.; Trnka, M.J.; Bushnell, D.A.; Davis, R.E.; Mattei, P.-J.; Burlingame, A.L.; Kornberg, R.D. Structure of a Complete Mediator-RNA Polymerase II Pre-Initiation Complex. Cell 2016, 166, 1411–1422e16.
- Cenik, B.K.; Shilatifard, A. COMPASS and SWI/SNF Complexes in Development and Disease. Nat. Rev. Genet. 2021, 22, 38–58.
- Tessarz, P.; Kouzarides, T. Histone Core Modifications Regulating Nucleosome Structure and Dynamics. Nat. Rev. Mol. Cell Biol. 2014, 15, 703–708.
- Boija, A.; Mahat, D.B.; Zare, A.; Holmqvist, P.-H.; Philip, P.; Meyers, D.J.; Cole, P.A.; Lis, J.T.; Stenberg, P.; Mannervik, M. CBP Regulates Recruitment and Release of Promoter-Proximal RNA Polymerase II. Mol. Cell 2017, 68, 491–503.e5.
- Levy, D. Lysine Methylation Signaling of Non-Histone Proteins in the Nucleus. Cell Mol. Life Sci. 2019, 76, 2873–2883.
- Nagasaka, M.; Miyajima, C.; Aoki, H.; Aoyama, M.; Morishita, D.; Inoue, Y.; Hayashi, H. Insights into Regulators of P53 Acetylation. Cells 2022, 11, 3825.
- Chuikov, S.; Kurash, J.K.; Wilson, J.R.; Xiao, B.; Justin, N.; Ivanov, G.S.; McKinney, K.; Tempst, P.; Prives, C.; Gamblin, S.J.; et al. Regulation of P53 Activity through Lysine Methylation. Nature 2004, 432, 353–360.
- Mannervik, M. Control of Drosophila Embryo Patterning by Transcriptional Co-Regulators. Exp. Cell Res. 2014, 321, 47–57.
- Kim, J.J.; Kingston, R.E. Context-Specific Polycomb Mechanisms in Development. Nat. Rev. Genet. 2022, 23, 680–695.
- Schuettengruber, B.; Chourrout, D.; Vervoort, M.; Leblanc, B.; Cavalli, G. Genome Regulation by Polycomb and Trithorax Proteins. Cell 2007, 128, 735–745.
- Kassis, J.A.; Kennison, J.A.; Tamkun, J.W. Polycomb and Trithorax Group Genes in Drosophila. Genetics 2017, 206, 1699–1725.
- Blackledge, N.P.; Klose, R.J. The Molecular Principles of Gene Regulation by Polycomb Repressive Complexes. Nat. Rev. Mol. Cell Biol. 2021, 22, 815–833.
- Vershinin, Z.; Feldman, M.; Werner, T.; Weil, L.E.; Kublanovsky, M.; Abaev-Schneiderman, E.; Sklarz, M.; Lam, E.Y.N.; Alasad, K.; Picaud, S.; et al. BRD4 Methylation by the Methyltransferase SETD6 Regulates Selective Transcription to Control MRNA Translation. Sci. Adv. 2021, 7, eabf5374.
- Erceg, J.; Pakozdi, T.; Marco-Ferreres, R.; Ghavi-Helm, Y.; Girardot, C.; Bracken, A.P.; Furlong, E.E.M. Dual Functionality of Cis-Regulatory Elements as Developmental Enhancers and Polycomb Response Elements. Genes Dev. 2017, 31, 590–602.
- Gisselbrecht, S.S.; Palagi, A.; Kurland, J.V.; Rogers, J.M.; Ozadam, H.; Zhan, Y.; Dekker, J.; Bulyk, M.L. Transcriptional Silencers in Drosophila Serve a Dual Role as Transcriptional Enhancers in Alternate Cellular Contexts. Mol. Cell 2020, 77, 324–337.e8.
- Huang, D.; Ovcharenko, I. Enhancer-Silencer Transitions in the Human Genome. Genome Res. 2022, 32, 437–448.
- Kuroda, M.I.; Kang, H.; De, S.; Kassis, J.A. Dynamic Competition of Polycomb and Trithorax in Transcriptional Programming. Annu. Rev. Biochem. 2020, 89, 235–253.
- De, S.; Cheng, Y.; Sun, M.-A.; Gehred, N.D.; Kassis, J.A. Structure and Function of an Ectopic Polycomb Chromatin Domain. Sci. Adv. 2019, 5, eaau9739.
- De, S.; Gehred, N.D.; Fujioka, M.; Chan, F.W.; Jaynes, J.B.; Kassis, J.A. Defining the Boundaries of Polycomb Domains in Drosophila. Genetics 2020, 216, 689–700.
- Bergman, D.T.; Jones, T.R.; Liu, V.; Ray, J.; Jagoda, E.; Siraj, L.; Kang, H.Y.; Nasser, J.; Kane, M.; Rios, A.; et al. Compatibility Rules of Human Enhancer and Promoter Sequences. Nature 2022, 607, 176–184.
- Martinez-Ara, M.; Comoglio, F.; van Arensbergen, J.; van Steensel, B. Systematic Analysis of Intrinsic Enhancer-Promoter Compatibility in the Mouse Genome. Mol. Cell 2022, 82, 2519–2531.e6.
- Maksimenko, O.; Golovnin, A.; Georgiev, P. Enhancer-Promoter Communication Is Regulated by Insulator Pairing in a Drosophila Model Bigenic Locus. Mol. Cell Biol. 2008, 28, 5469–5477.
- Savitskaya, E.; Melnikova, L.; Kostuchenko, M.; Kravchenko, E.; Pomerantseva, E.; Boikova, T.; Chetverina, D.; Parshikov, A.; Zobacheva, P.; Gracheva, E.; et al. Study of Long-Distance Functional Interactions between Su(Hw) Insulators That Can Regulate Enhancer-Promoter Communication in Drosophila Melanogaster. Mol. Cell Biol. 2006, 26, 754–761.
- Batut, P.J.; Bing, X.Y.; Sisco, Z.; Raimundo, J.; Levo, M.; Levine, M.S. Genome Organization Controls Transcriptional Dynamics during Development. Science 2022, 375, 566–570.
- Levo, M.; Raimundo, J.; Bing, X.Y.; Sisco, Z.; Batut, P.J.; Ryabichko, S.; Gregor, T.; Levine, M.S. Transcriptional Coupling of Distant Regulatory Genes in Living Embryos. Nature 2022, 605, 754–760.
- Aljahani, A.; Hua, P.; Karpinska, M.A.; Quililan, K.; Davies, J.O.J.; Oudelaar, A.M. Analysis of Sub-Kilobase Chromatin Topology Reveals Nano-Scale Regulatory Interactions with Variable Dependence on Cohesin and CTCF. Nat. Commun. 2022, 13, 2139.
- Lim, B.; Levine, M.S. Enhancer-Promoter Communication: Hubs or Loops? Curr. Opin. Genet. Dev. 2021, 67, 5–9.
- Karr, J.P.; Ferrie, J.J.; Tjian, R.; Darzacq, X. The Transcription Factor Activity Gradient (TAG) Model: Contemplating a Contact-Independent Mechanism for Enhancer-Promoter Communication. Genes Dev. 2022, 36, 7–16.
- Fujioka, M.; Jaynes, J.B.; Goto, T. Early Even-Skipped Stripes Act as Morphogenetic Gradients at the Single Cell Level to Establish Engrailed Expression. Development 1995, 121, 4371–4382.
- Fujioka, M.; Emi-Sarker, Y.; Yusibova, G.L.; Goto, T.; Jaynes, J.B. Analysis of an Even-Skipped Rescue Transgene Reveals Both Composite and Discrete Neuronal and Early Blastoderm Enhancers, and Multi-Stripe Positioning by Gap Gene Repressor Gradients. Development 1999, 126, 2527–2538.
- Sackerson, C.; Fujioka, M.; Goto, T. The Even-Skipped Locus Is Contained in a 16-Kb Chromatin Domain. Dev. Biol. 1999, 211, 39–52.
- Small, S.; Blair, A.; Levine, M. Regulation of Two Pair-Rule Stripes by a Single Enhancer in the Drosophila Embryo. Dev. Biol. 1996, 175, 314–324.
- Frasch, M.; Hoey, T.; Rushlow, C.; Doyle, H.; Levine, M. Characterization and Localization of the Even-Skipped Protein of Drosophila. EMBO J. 1987, 6, 749–759.
- Macdonald, P.M.; Ingham, P.; Struhl, G. Isolation, Structure, and Expression of Even-Skipped: A Second Pair-Rule Gene of Drosophila Containing a Homeo Box. Cell 1986, 47, 721–734.
- Peel, A.D.; Chipman, A.D.; Akam, M. Arthropod Segmentation: Beyond the Drosophila Paradigm. Nat. Rev. Genet. 2005, 6, 905–916.
- Clyde, D.E.; Corado, M.S.G.; Wu, X.; Paré, A.; Papatsenko, D.; Small, S. A Self-Organizing System of Repressor Gradients Establishes Segmental Complexity in Drosophila. Nature 2003, 426, 849–853.
- Pankratz, M.J.; Jäckle, H. Making Stripes in the Drosophila Embryo. Trends Genet. 1990, 6, 287–292.
- Pankratz, M.J.; Seifert, E.; Gerwin, N.; Billi, B.; Nauber, U.; Jäckle, H. Gradients of Krüppel and Knirps Gene Products Direct Pair-Rule Gene Stripe Patterning in the Posterior Region of the Drosophila Embryo. Cell 1990, 61, 309–317.
- Struhl, G.; Johnston, P.; Lawrence, P.A. Control of Drosophila Body Pattern by the Hunchback Morphogen Gradient. Cell 1992, 69, 237–249.
- Martinez-Arias, A.; Lawrence, P.A. Parasegments and Compartments in the Drosophila Embryo. Nature 1985, 313, 639–642.
- Small, S.; Kraut, R.; Hoey, T.; Warrior, R.; Levine, M. Transcriptional Regulation of a Pair-Rule Stripe in Drosophila. Genes Dev. 1991, 5, 827–839.
- Lim, B.; Fukaya, T.; Heist, T.; Levine, M. Temporal Dynamics of Pair-Rule Stripes in Living Drosophila Embryos. Proc. Natl. Acad. Sci. USA 2018, 115, 8376–8381.
- Small, S.; Arnosti, D.N. Transcriptional Enhancers in Drosophila. Genetics 2020, 216, 1–26.
- Liang, H.-L.; Nien, C.-Y.; Liu, H.-Y.; Metzstein, M.M.; Kirov, N.; Rushlow, C. The Zinc-Finger Protein Zelda Is a Key Activator of the Early Zygotic Genome in Drosophila. Nature 2008, 456, 400–403.
- Tsurumi, A.; Xia, F.; Li, J.; Larson, K.; LaFrance, R.; Li, W.X. STAT Is an Essential Activator of the Zygotic Genome in the Early Drosophila Embryo. PLoS Genet. 2011, 7, e1002086.
- Struffi, P.; Corado, M.; Kaplan, L.; Yu, D.; Rushlow, C.; Small, S. Combinatorial Activation and Concentration-Dependent Repression of the Drosophila Even Skipped Stripe 3 + 7 Enhancer. Development 2011, 138, 4291–4299.
- Vincent, B.J.; Staller, M.V.; Lopez-Rivera, F.; Bragdon, M.D.J.; Pym, E.C.G.; Biette, K.M.; Wunderlich, Z.; Harden, T.T.; Estrada, J.; DePace, A.H. Hunchback Is Counter-Repressed to Regulate Even-Skipped Stripe 2 Expression in Drosophila Embryos. PLoS Genet. 2018, 14, e1007644.
- Mir, M.; Stadler, M.R.; Ortiz, S.A.; Hannon, C.E.; Harrison, M.M.; Darzacq, X.; Eisen, M.B. Dynamic Multifactor Hubs Interact Transiently with Sites of Active Transcription in Drosophila Embryos. eLife 2018, 7, e40497.
- Fujioka, M.; Sun, G.; Jaynes, J.B. The Drosophila Eve Insulator Homie Promotes Eve Expression and Protects the Adjacent Gene from Repression by Polycomb Spreading. PLoS Genet. 2013, 9, e1003883.
- Fujioka, M.; Wu, X.; Jaynes, J.B. A Chromatin Insulator Mediates Transgene Homing and Very Long-Range Enhancer-Promoter Communication. Development 2009, 136, 3077–3087.
- Fujioka, M.; Mistry, H.; Schedl, P.; Jaynes, J.B. Determinants of Chromosome Architecture: Insulator Pairing in Cis and in Trans. PLoS Genet. 2016, 12, e1005889.
- Baxley, R.M.; Bullard, J.D.; Klein, M.W.; Fell, A.G.; Morales-Rosado, J.A.; Duan, T.; Geyer, P.K. Deciphering the DNA Code for the Function of the Drosophila Polydactyl Zinc Finger Protein Suppressor of Hairy-Wing. Nucleic Acids Res. 2017, 45, 4463–4478.
- Cuartero, S.; Fresán, U.; Reina, O.; Planet, E.; Espinàs, M.L. Ibf1 and Ibf2 Are Novel CP190-Interacting Proteins Required for Insulator Function. EMBO J. 2014, 33, 637–647.
- Melnikova, L.; Kostyuchenko, M.; Molodina, V.; Parshikov, A.; Georgiev, P.; Golovnin, A. Multiple Interactions Are Involved in a Highly Specific Association of the Mod(Mdg4)-67.2 Isoform with the Su(Hw) Sites in Drosophila. Open Biol. 2017, 7, 170150.
- Maksimenko, O.; Bartkuhn, M.; Stakhov, V.; Herold, M.; Zolotarev, N.; Jox, T.; Buxa, M.K.; Kirsch, R.; Bonchuk, A.; Fedotova, A.; et al. Two New Insulator Proteins, Pita and ZIPIC, Target CP190 to Chromatin. Genome Res. 2015, 25, 89–99.
This entry is offline, you can click here to edit this entry!
