The multicellular volvocine species are included in three families: Tetrabaenaceae, Goniaceae, and Volvocaceae. In addition, within the Volvocaceae family, two sub-clades have been defined: the “Eudorina Group” and the Euvolvox (or section Volvox) [
29] (
Figure 1). The Tetrabaenaceae family contains two species,
Tetrabaena socialis and
Basichlamys sacculifera. These are the simplest multicellular volvocine algae with four
Chlamydomonas-like cells arranged like a four-leaf clover [
30,
31]. The polyphyletic Goniaceae family includes several species in two genera,
Gonium and
Astrephomene. The
Gonium species have 8–16
Chlamydomonas-like cells arranged as flat plates, whereas the
Astrephomene species are 32- or 64-celled spheroidal colonies with 2 to 4 sterile somatic cells in the posterior of the colony [
26,
32,
33]. The Volvocaceae is the largest and most diverse family of volvocine green algae with many polyphyletic genera. Algae in the genera
Eudorina, Pandorina, Volvulina, Yamagishiella, and
Colemanosphaera all have spheroidal body plans with between 16 and 64 cells (cell numbers vary between genera) with no germ–soma cellular differentiation under standard growth conditions. Species in the
Pleodorina genus have 32 to 128 cells with specialized somatic cells in the anterior portion of the colony, except for one species,
Pleodorina sphaerica, that has somatic cells distributed in both the anterior and posterior of the colony [
26,
34]. Finally, species in the genus
Volvox are the largest and most complex members of the Volvocaceae, with several hundred to several thousand cells and two distinct cell types, specialized germ and specialized soma [
26].
The species
V. carteri forma
nagariensis has served as the primary model organism for studying the developmental and genetic mechanisms that underlie cellular differentiation [
23,
36,
37]. An asexual
V. carteri individual contains 1000–2000 small
Chlamydomonas-like flagellated somatic cells and up to 16 large unflagellated germ cells known as gonidia. The somatic cells are terminally differentiated and have no cell division potential. The lack of cell division ensures that the motility of the individual is maintained because flagellar activity is compromised during cell division in volvocine algae due to the so-called “flagellation constraint” and the presence of a rigid cell wall [
38]. Juvenile
V. carteri develop from gonidia, which grow to up to ~1000 times the volume of somatic cells before they start dividing. The development of a juvenile
V. carteri begins with a series of 5 symmetric divisions resulting in a 32-celled embryo. Then, during the sixth division cycle, the sixteen cells in the anterior of the embryo divide asymmetrically with one daughter cell inheriting a larger volume of cytoplasm than the other. These large cells go through 2 additional asymmetric divisions and then cease dividing, while all other cells go through a series of 11 to 12 symmetric divisions. At the end of cleavage, the embryo contains ~2000 cells, most of which are small soma initial cells, except the 16 large germ cell initials generated through asymmetric division. At this stage, the embryo is effectively inside-out relative to the adult organization, with the flagella of the somatic cells pointing inward. To gain the adult configuration, the embryo goes through an inversion process.
2. regA Gene Structure and Function
Early investigations into cellular differentiation in
V. carteri identified a class of mutants called “somatic regenerators” in which somatic cells appear to first develop normally but then dedifferentiate and become reproductive [
44,
45]. Linkage analysis found that all such regenerator mutants map onto a single locus which was named
regA (from “
regenerator”) [
36,
46,
47]. Huskey & Griffin [
47] originally described a second
regB locus based on linkage group analysis of regenerator mutants, but reexamination of
regB mutants by members of the same research lab determined that they are not regenerator mutants and have a different mutant phenotype [
48]. Thus, in retrospect, all regenerator mutants can be mapped onto the
regA locus [
36]. However, it is worth noting that the annotation “RegA” or “Reg genes” has been used multiple times independently in species from other groups (e.g., bacteria and animals) to refer to different genes coding for distinct unrelated proteins. Such similarities in name are due to historical and linguistic coincidence rather than any shared function or homology. In this review, we are strictly discussing the
regA gene and its gene family that is restricted to volvocine algae.
Based on the link between somatic regeneration and the
regA locus, the
regA gene was deemed the master regulatory gene that controls somatic cell development in
V. carteri [
36,
37,
49]. Kirk et al. [
49] used transposon tagging to identify the
regA gene and went on to determine that the RegA protein is localized in the nuclei of somatic cells. In
V. carteri f.
nagariensis,
regA is expressed exclusively in somatic progenitor cells, with its transcription beginning early in development shortly after inversion [
49,
50,
51,
52].
regA transcript levels appear to persist and fluctuate throughout the life cycle [
49], but see the study by König and Nedelcu [
24] for an alternative possibility and discussion.
The functional role of RegA, its amino acid composition, and the presence of a DNA-binding SAND domain in the RegA protein [
53] helped establish the current working model in which RegA acts as a transcriptional repressor of genes needed for gonidial development [
37]. A long-standing hypothesis is that
regA suppresses the expression of nuclear-encoded chloroplast proteins required for chloroplast biogenesis and turn-over [
54,
55,
56]. These negative effects on the chloroplasts would be reflected in the inability of the somatic cells to photosynthesize, grow, and divide. However, Matt and Umen [
52] cast some doubt on this idea. They used whole transcriptome analysis to compare the expression profiles of germ cells and somatic cells. While photosynthetic genes were expressed at around two-fold higher levels in germ cells, photosynthetic genes were nevertheless highly abundant in somatic cells as well. Matt and Umen [
52] propose that both germ cells and somatic cells maintain active photosynthesis, but germ cells are specialized in anabolic processes such as starch, fatty acid, and amino acid biosynthesis, while somatic cells break down starch and lipids to provide the substrates needed to synthesize ECM glycoproteins. Therefore, while it remains plausible that
regA downregulates photosynthetic genes, it is also possible that
regA downregulates other genes related to germ cell growth such as starch synthesis.
The structure of the
regA gene has been well described for
V. carteri and serves as the basic template for the gene structures of many other homologs of
regA in the VARL (
volvocine
algae
regA-like) gene family. The minimal promoter of
regA consists of only 42 nt found directly upstream of the transcription start site with a plausible TATA box with the sequence TAATTGA beginning at −28 and an initiator region with the sequence CACTCAT beginning -1 relative to the transcription start site [
57]. The transcriptional unit of
regA is 12,477 nt long and contains 7 introns and 8 exons. After the introns are spliced out, the mature
regA mRNA is 6725 nt long and consists of a 940 nt 5′UTR (exons 1–5), a 3147 nt coding region (exons 5–8), and a 2638 nt 3′UTR with a UGUAA polyadenylation signal [
49].
However, a splice variant that retains intron 7 (1194 bp) is expressed at low levels in
V. carteri f.
nagariensis as well. The donor splice site of intron 7 is GC instead of the typical GU, which may explain the variation in splicing. Remarkably, intron 7 encodes an ORF in the same frame as the rest of the
regA coding region and, therefore, is likely to be translated, resulting in two different RegA protein products. However, experiments using modified
regA transformation constructs to alter the splicing and translation of intron 7 have demonstrated that the presence or absence of intron 7 splicing has no detectable effect on the phenotypic rescue of regenerator mutants, despite the retention of intron 7 adding nearly 400 more amino acid residues to the RegA protein [
57]. Interestingly, the homologous region to intron 7 is not spliced out in the closely related
V. carteri f.
kawasakiensis, and protein-level homology has been described in the intron 7 region across a wide variety of volvocine algae species [
25,
53]. Thus, it appears likely that splicing out intron 7 is a quirk specific to
V. carteri f.
nagariensis, while homologous regions are exonic in other species.
In addition to the promoter, the differential transcription of
regA is regulated by two enhancers found in introns 3 and 5 and a silencer found in intron 7 [
57]. Eight possible AUG start codons are found in the 5′UTR of mature
regA mRNA and are thought to be bypassed via a ribosome shunting mechanism so that translation begins at the ninth AUG sequence of the mRNA [
58].
Following translation, the predicted RegA protein is 1049-amino-acids-long without the inclusion of intron 7 or 1447 with intron 7 and contains a high proportion of glutamine, alanine, and proline residues [
49,
57]. A key structural region within the RegA protein is the VARL domain, which is the distinguishing feature of the VARL gene family [
53,
59]. The VARL domain is located between amino acids 444 and 558 in the RegA of
V. carteri f.
nagariensis and is composed of a highly conserved core VARL region (sites 484–558), a short but highly conserved N-terminal extension region (sites 444–455), and a less conserved linker region between these two [
25,
53,
59]. In addition, two short motifs of high amino acid conservation have been identified that are shared across the predicted RegA proteins of numerous volvocine algae species: a “LALRP” motif upstream of the VARL domain and an “FLQ” motif found within the intron 7 region downstream of the VARL domain [
25].
The core VARL domain appears to encode a DNA-binding SAND domain [
53]. The SAND domain (IPR000770/PF01342)—named after
Sp100,
AIRE-1,
NucP41/75, and
DEAF-1—is a DNA-binding domain found in animal and plant proteins that function in chromatin-dependent transcriptional control or bind-specific DNA sequences (e.g., [
60]). SAND-containing proteins are involved in multiple distinct processes, both general and lineage/tissue-specific. However, most of the SAND-containing proteins with known functions are involved in multicellular development, including cell differentiation, cell proliferation, tissue homeostasis, and organ formation. For instance, DEAF-1 (
Deformed
Epidermal
Autoregulatory
Factor-
1) is involved in breast epithelial cell differentiation in mammals [
61] and is necessary for embryonic development in
Drosophila melanogaster [
62]. GMEB (
Glucocorticoid
Modulatory
Element
Binding) regulates neural apoptosis in the nematode
Caenorhabditis elegans [
63]. Spe44 (
Speckled protein
44 kDa) is a master switch for germ cell fate in
C. elegans and, like the mammalian AIRE1 (
Auto
immune
Regulator
1), plays a role in sperm cell differentiation [
64,
65,
66]. In land plants, SAND domains are associated with ATX (the
Arabidopsis homolog of
trithora
x) and ULTRAPETALA (ULT) proteins, which are involved in cell proliferation, cell differentiation, and tissue patterning. Specifically, ATX1 in
Arabidopsis thaliana is required for root, leaf, and floral development through its histone methyltransferase activity [
67], and ULT is a negative regulator that influences shoot and floral meristem size by controlling cell accumulation [
68,
69,
70].
3. regA-like Gene Family Evolution
The VARL gene family is defined by the presence of a homologous VARL domain within the predicted protein (note that volvocine algae possess additional SAND-containing proteins outside the VARL family). Although all VARL genes contain the VARL domain, the sequence level conservation outside of the VARL domain is very low. Thus, entire gene sequences cannot be aligned and used for phylogenetic analyses. The VARL domain itself is very short (~86 amino acids) and not highly conserved, such that its utility for inferring evolutionary relationships between the members of the VARL gene family is also limited. Nevertheless, information from gene synteny, sequence signatures outside of the VARL domain, and the locations of conserved introns can help draw more robust conclusions regarding the evolution of the VARL family [25].
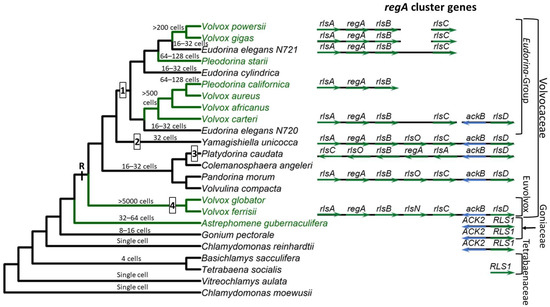
Based on currently available whole genome sequence data, the VARL gene family contains 12 members in C. reinhardtii [59], 8 in G. pectorale [32] and T. socialis [72], 6 in A. gubernaculifera [33], and 14 in V. carteri [59]. With the exception of regA orthologs (when present), all other regA homologs are known as regA-like sequences, annotated as RLS1-12 in Chlamydomonas and Goniaceae or rlsA-O in Volvocaceae. C. reinhardtii and other volvocine algae outside the Volvocaceae lack orthologs of any of the regA cluster genes. The closest homolog to the regA cluster genes found in these species is RLS1. This gene is an ortholog of the Volvocaceaen rlsD, which is the closest rls paralog of the regA cluster. Currently it is thought that the VARL gene family comprising several paralogs including RLS1/rlsD was already present in the common ancestor of all volvocine green algae. RLS1/rlsD underwent one or more duplication events in the common ancestor of the Volvocaceae family to give rise to a five-gene regA gene cluster comprising rlsA, regA, rlsB, rlsO, and rlsC. After the lineage leading to V. ferrisii diverged from the rest of the Volvocaceae, its rlsO gene gained a second VARL domain and evolved into rlsN. Meanwhile, the common ancestor of the Eudorina group lost rlsO. In addition, Y. unicocca lost two internal regA cluster genes (regA, rlsB, or rlsO) but restored the five-gene cluster via gene duplication, and the regA cluster of P. caudata became inverted relative to nearby syntenic markers (Figure 1).
Based on its role in suppressing reproduction in somatic cells, it has been hypothesized that regA evolved from a gene that was involved in trading off reproduction for survival (i.e., a life history trade-off gene) in the single-celled ancestors of V. carteri. Specifically, such a gene could have been co-opted by changing its expression from a temporal context (in response to an environmental cue) into a spatial context (in response to a developmental cue) [17].The common ancestor of V. carteri and C. reinhardtii likely had several VARL gene family members, one of which was RLS1. The RLS1 gene duplicated several times to give rise to the regA gene cluster in the common ancestor of the Volvocaceae, setting the stage for the functional co-option of regA during the evolution of cellular differentiation as well as other lineage-specific changes to regA cluster genes (Figure 1). The co-option of RLS1′s functions into a regA-like gene responsible for somatic cell differentiation likely involved the simulation of the ancestral environmentally induced signal in a developmental context.