1. General Concepts
miRNAs are sncRNAs with a length of 20–24 nucleotides that control gene expression posttranscriptionally [
5]. A distinct miRNA species acts by regulating one or multiple transcripts at the same time (up to hundreds), and a single mRNA species often exhibits multiple interaction sites for various miRNAs, thus generating complex regulatory circuits. As a result, even while single miRNAs commonly have a restrained effect on a particular target, their action produces cumulative effects by negatively influencing different transcripts in a signalling network. Accordingly, metazoan miRNAs have been dubbed the sculptors of the cell transcriptome [
6].
These short molecules work as gene silencers by way of base-pairing to a target mRNA in the 3′ untranslated sequence even though they have been observed to bind anywhere alongside the mRNA sequence. Interestingly, miRNAs can act either by influencing the stability and degradation of the target mRNA or by suppressing its translation. Because they are essential for base pairing with a target mRNA, the nucleotides at positions 2–8 of a mature miRNA have been termed a "seed sequence"; however, a fraction of miRNA-target recognition is also mediated through noncanonical seed-like consensus sequences. In addition, seed-region identity has also been used to classify miRNAs into families that have a common capacity to recognize sets of target mRNAs [
7].
Currently, 3012 unique human miRNAs have been recognized and stored in the miRTarBase repository, with a huge, estimated capacity of 4,475,477 potential miRNA–target interactions [
8]. Furthermore, several issues have been reported regarding the quality of miRNA annotations in publicly available databases, as well as the reproducibility of microRNA studies [
9]. Comprehensively, endogenous miRNAs have been reported to regulate most, if not all, protein-coding genes in mammals. However, with hundreds of conceivably miRNA–mRNA interactions, the human miRNA targetome is far from being determined [
10].
The miRNA biogenesis pathway is accountable for the handling of a pre-miRNA to a mature miRNA and has been widely reported [
11,
12,
13]. Of note, miRNAs are themselves sensitive to transcriptional and posttranscriptional control, and emerging approaches for the direct mapping of RNA modifications in sncRNAs have been proposed, with an emphasis on mass spectrometry and nanopore technologies [
14]. As an example, the terminal nucleotidyltransferases (rNTrs) can modulate miRNA sequences by adding nontemplated nucleotides to the 3’-end (tailing). Accordingly, miRNA isoforms with diverse 3′-ends (and possibly functions), known as 3′ isomiRs, have been thoroughly detected by NGS [
15]. The editing by adenosine deaminase acting on RNA (ADAR) is a relevant regulatory mechanism acting at the posttranscriptional level and is increasingly found in metazoans [
16]. The enzyme induces A-to-I RNA editing modification of double-stranded RNA. Editing leads to changes in miRNA processing, as well as the editing of a target mRNA, thus upsetting the mutual complementarity and driving a microRNA–mRNA interactional redirection, distressing diverse cellular processes. Of note, the editing activity of ADAR is increasingly reported to be enhanced in cancer, but its influence on deriving RNA modifications is still unknown [
16].
Excitingly, several endogenous cellular RNAs, such as circular RNAs, long noncoding RNAs, transcribed pseudogenes, and mRNAs, act as natural miRNA sponges, able to firmly interact with and disrupt target miRNAs, thus distressing their controlled networks [
17].
Although a small number of miRNAs have a tissue-specific distribution, most reveal a broad tissue localization. Given the pervasive presence of nucleases, it is assumed that RNA molecules are prone to rapid degradation in the extracellular environment. Nevertheless, miRNA molecules have been found to retain unexpected stability in plasma and other physiological fluids since they can be packaged from cells in the form of microvesicles and exosomes or protein complexes, such as Argonaute 2 (AGO-2) (taking part in the RNA silencing complex), nucleophosmin-1 (NPM-1) (an RNA-binding protein involved in ribosome nuclear export), or high-density lipoproteins. These forms are able to be actively released into the extracellular space and reach recipient cells, where they can influence the translation of target genes, thus defining an explicit role of miRNAs in cell–cell communication [
18]. Accordingly, the discovery of about 300 circulating miRNAs (c-miRNAs) has highlighted miRNAs’ potential as intercellular signalling molecules and disease biomarkers [
19] since a dysregulated expression of miRNA can unsettle tightly controlled RNA networks in tissues and organs, potentially contributing to abnormalities [
20]. Nevertheless, most of the functional significance of c-miRNAs remains unknown to date, and the gap between discovery and function has yet to be bridged.
Many studies have found correlations between altered levels of miRNA and the physiopathology of numerous processes including mitochondrial [
21], cardiovascular [
22], neurodegenerative [
23], immune disease [
24], inflammatory [
25], rare genetic disorders, and more; most of these studies have focused on miRNAs as biomarkers of cancer [
26,
27,
28,
29,
30], emphasizing their relevance as personalized theranostic factors. Interestingly, miRNAs are not only prospective markers of the onset and progression of neoplastic disease, but also indicators of responses to cancer therapy, as well as their relationship to drug resistance and the modulation of responses to cancer treatment [
31].
By controlling crucial metabolic processes, miRNAs have also been found to play a role in energy balance and the oversight of metabolic pathways in living organisms [
20,
32,
33,
34,
35,
36,
37,
38]. Furthermore, the treatment of infectious diseases has been linked to several miRNAs [
39]. Yet, it has been discovered that miRNA expression changes in response to dietary and lifestyle factors [
40]; consequently, the recent nutrimiRomics research focuses on the impact of diet on miRNA levels in the human body, as well as their downstream impact on gene expression and subsequent health outcomes. Finally, there is growing evidence that dietary miRNAs may survive digestion [
41]. However, the roles and capabilities of food-related miRNAs in modulating interspecies RNA are still an enigma.
2. Technological Advances Offer New Opportunities for miRNAs in Translational Medicine
Since the first report of a noncoding RNA identified in
C. elegans in 1993 [
42], the bulk of the 153,210 papers on miRNA research listed in PubMed (January 2023) reveals their critical impact on human diseases. The substantial translational interest is confirmed by searching Google’s patent database; a search for the keywords “microRNA” and “biomarker” yielded 56,021 results (as of January 2023). The quantity and diversity of studies are evocative of the astonishing complexity and limitations of miRNA research that is increasingly projected toward next-generation medicine. According to the unusual levels of miRNAs found in some unhealthy conditions, novel molecular diagnostic strategies are expected, deeply encouraging researchers and producers to focus on miRNAs as relevant noninvasive disease biomarkers. However, so far, there is no agreement on specific miRNAs suitable for early disease detection, even in crucial topic areas such as in vivo cancer research [
43]. Current limitations in this discovery field are related to c-miRNA recognition methods, which must be extremely specific and able to recognize small quantities of target molecules, while also taking into account the presence of unwanted contaminants and inhibitors that could interfere with downstream analytical methodologies. Differences in all the critical steps used for miRNA extraction, detection, data normalization, validation, target gene identification, and experimental design are responsible for at least some of the discrepancies between the different studies [
44]. Overall, these constraints require the timely development of standard procedures and well-recognized guidelines to accurately decipher the power and versatility of miRNAs in molecular diagnostics [
45,
46].
Furthermore, the prompt increase of RNA drugs in recent research and clinical development, driven in part by the success of messenger RNA vaccines in the fight against the SARS-CoV-2 pandemic, has spurred the pursuit of mRNA-based drugs for treating other conditions.
Therapeutic agents are classically based on synthetic small molecules, monoclonal antibodies, or large proteins. Traditional drugs may fail to hit intended therapeutic targets due to the inaccessibility of active sites in the target’s three-dimensional structure. RNA-based therapies can offer an exceptional opportunity to potentially reach any relevant target with therapeutic purposes. Additionally, techniques based on nucleic acid technologies can avoid the need for laborious synthesis procedures. Yet, the recognition sequence can be quickly revised and adapted to the target. Nevertheless, RNA-based therapies have specificity problems that carry the risk of side effects [
47,
48]; in addition, RNA-based drugs’ susceptibility to degradation may determine poor pharmacodynamics [
47,
48]. A number of these issues can be mitigated by chemical changes in the structure of the synthetic polymer (RNA or DNA). Of note, RNA-based drugs are usually larger in size than small-molecule therapies and carry electrical charges, which makes their intracellular delivery in their native form difficult.
In this context, the therapeutic potential of miRNA treatments is receiving attention in clinical trials of almost all human diseases [
43,
49]. Given the ability of miRNAs to target specific mRNAs, inhibitors based on the sequence of overexpressed miRNAs can be used prospectively in the form of anti-miRs to lower elevated levels of miRNAs and, in turn, restore their downregulated transcripts [
50]. This opportunity for medically-based interventions is closely linked to the pioneering use of antisense molecules, the first molecular drugs inspired by the target sequence [
51]. These synthetic tools have a sequence complementary to a specific mRNA whose levels they can modulate [
52]. Anti-miRs are typically based on first-generation antisense single-stranded oligonucleotides (ASOs), or their chemically modified forms as blocked nucleic acids (LNAs), which are opportunely designed to recognize target mRNAs. Yet, anti-miRs with a 2ʹ-O-methoxyethyl substitution are classified as antagomiRs [
53]. Furthermore, miRNA mimics are chemically synthesized, double-stranded small RNA molecules which mimic mature endogenous miRNAs (miRNA replacement therapy) after transfection into cells, the action of which is aimed at replacing downregulated or missing miRNA expression [
54].
The delivery of virus-mediated miRNA-based therapies has shown great success in animal models, where adenoviruses have been successful in delivering both anti-miR and miRNA mimics. However, despite the high efficiency of virus-based miRNA delivery systems, several concerns continue, including toxicity, immunogenicity, and insert-size constraints [
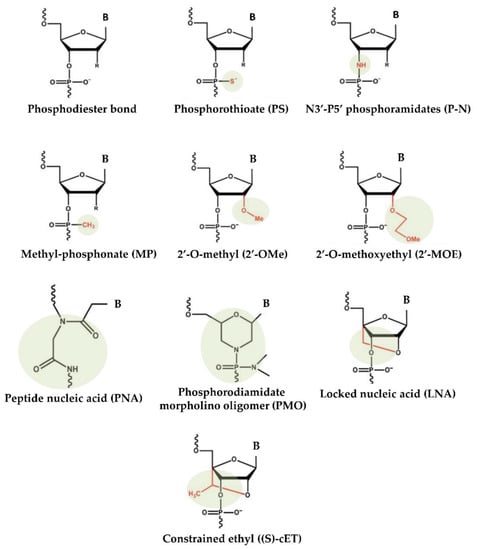
55]. To overcome these difficulties, over the years, nonviral approaches for the production and delivery of miRNA-based drugs have emerged. Substantial improvements in designing, synthesis, binding affinity, stability, and target modulation effects of both miRNA mimics and anti-miRs have been accomplished through chemical changes to the nucleotide backbone since a major challenge for RNA-based therapeutic strategies is the possible degradation of oligonucleotides by RNases in extracellular and endocellular compartments. Accordingly, the oligonucleotide chemistry has been widely altered by modifying the nucleotides, introducing base modifications, modifications on ribose moiety, and modifications to the phosphate group in the sugar−phosphate backbone (
Figure 1). As an example, by introducing methylation (2ʹ-O-methoxyethyl), locked nucleic acids (LNAs), a nucleic acid analogue that contains a 2′-O-methoxyethyl, 2′-fluoro and 2′,4′-methylene bridge, peptide nucleic acid (PNA) a synthetic polymer, an analogue to DNA and RNA, in which the sugar–phosphate backbone has been substituted by a unit of N-(2-aminoethyl) glycine, adding phosphorothioate (PS)-like groups (by replacing the nonbridging oxygen in the phosphate group with sulphur) [
51], and others [
56]. Overall, these changes have been shown to increase the stability of oligonucleotides by making internucleotide linkages more resistant to the degradation of nucleases, and preserving both the ability to activate RNase H (the enzyme responsible for mRNA target cleavage) and their function in suppressing the target gene. In this research field, the diverse anti-miRNA oligonucleotide chemistries were also directly compared, and the in vitro results showed that the combination of 2′-O-methyl and LNA with phosphorothioate ends was about 10 times more active than modification alone, and 2 times as effective as the 2′-O-methyl with LNA changes [
57]. Overall, up to now, most efforts to develop therapies based on chemically modified miRNAs have been directed toward the development of anti-miRs using LNA chemistry, whereas currently commercially available miRNA mimics are often modified by methylation [
58,
59].
Figure 1. Chemical changes of oligonucleotides used in main clinical trials.
Another strategic approach facilitating RNA-based therapeutics in clinical application consists of encapsulating miRNA mimics or anti-miRs in carrying vehicles to confer to them protection from nuclease.
Therapeutic miRNAs include negatively charged polymers that cannot directly cross cell membranes. To get to their intended targets, they need appropriate formulations, including carriers as well as chemical modifications. Accordingly, delivery systems must shield the therapeutic RNAs from serum nucleases, avoid immune-system interference, escape unintended interactions with serum proteins, and prevent renal clearance when given systemically. Previous studies have shown that therapeutic RNAs administered locally or topically can circumvent complications associated with systemic administration, have higher bioavailability, and have a reduced clearance as compared to those administered systemically; nevertheless, this delivery opportunity is restricted to tumours, mucous membranes, eyes, and skin [
60]. In this context, the subcutaneous administration of naked mRNA, as compared with mRNA-loaded nanoparticles, leads to a more effective translation of the protein [
61].
The nanocarrier encapsulation strategy can both protect and deliver the drug to recipient cells, while biological obstacles, such as immunogenicity and nuclease, are often approached by chemically modifying the nucleotide structure [
62]. Various transport methods are increasingly being used to improve bioavailability, including liposomes and biodegradable polymers. In this context, liposomes, long used as immunological adjuvants in vaccination, adequately fulfil this role [
63].
Lipid nanoparticles are the most commonly used nonviral delivery technology for nucleic-acid-based drugs and vaccines [
64]. They are composed of complexes of anionic nucleic acids and synthetic cationic lipids. The cationic lipids characteristically include amine derivatives, such as primary, secondary, and tertiary amines, in addition to quaternary ammonium, amidinium salts, and various amine combinations [
65]. Furthermore, guanidine, imidazole groups (pyridinium, piperazine), and amino acids, including tryptophan, lysine, arginine, and ornithine, have also been employed [
66]. A comprehensive list of commonly used cationic lipids in drug formulations was recently reviewed [
66]. The advantages of lipid-based transfer systems consist of ease of assembly, biodegradability, the ability to shield nucleic acids entrapped by nucleases, the protection of renal clearance, the ability to promote cellular uptake, and the avoidance of endosomes. Additional carriers with biodegradability, biocompatibility, and low toxicity are employed. Polymeric nanoparticles are among the most-studied carriers and include synthetic and natural cationic polymers, as well as polyethylenimine (PEI), cyclodextrin, chitosan, and poly (lactic-co-glycolic acid) (PLGA) [
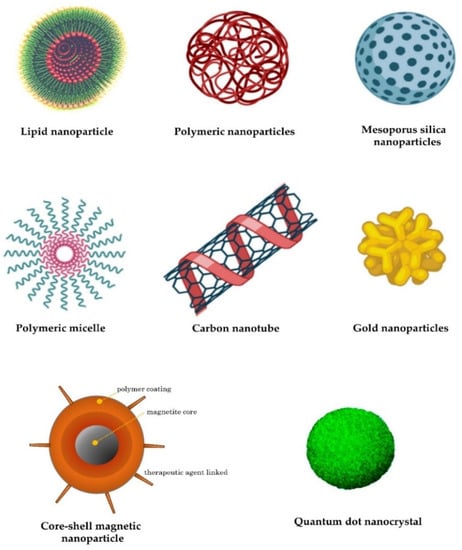
67]. Furthermore, dendrimers, carbon nanotubes, peptides, gold nanoparticles, silica-derived nanoparticles, iron-oxide-based magnetic nanoparticles, and other nanoparticles are increasingly being studied for medical interventions (
Figure 2) [
62].
Figure 2. Examples of nanocarriers as drug-delivery systems.
This entry is adapted from the peer-reviewed paper 10.3390/genes14020314