RUNX2 RNA and protein expression levels in various types of cancer were measured. Relatively high RUNX2 levels were detected by IHC staining in tissues of renal cell carcinoma compared with nontumor tissue, whose regulatory mechanism required Zic family member 2 (Zic2) in 786-O and ACHN cells [
12]. RUNX2 was shown to be an interactive target of miR-23a-3p in CAL-27 cells and TSCCA cells, and oral squamous cell carcinoma (OSCC) overexpressing miR-23a-3p mimics decreased the RUNX2 level [
15]. RUNX2 was significantly decreased by transfection of a miRNA-218 mimic, and RUNX2 expression was obviously increased by treatment with a miRNA-218 inhibitor in osteosarcoma U2OS cells [
20]. In oral cancer (both HSC-3 and Ca9-22 cells), RUNX2 expression was positively regulated by MRE11, the nuclease component of the RAD50/MRE11/NBS1 DNA repair complex [
57]. In a colorectal cancer study that enrolled 75 cancer patients, cancer tissues displayed high RUNX2 levels compared with normal adjacent tissues. Consistent results with these were obtained by Western blot analysis of 10 paired cancer and normal tissues [
17]. RUNX2 protein was detected in cervical cancer tissues, and RUNX2 expression declined upon overexpression of miR-218-5p in C-33A and CaSki cells [
58]. RUNX2 protein was elevated in human thyroid cancer cell lines and cancer tissues compared with primary cell lines and normal thyroid tissues [
10]. RUNX2 was overexpressed in lung adenocarcinoma in a large study that included 2418 tumor and 1574 nontumor lung samples [
59]. In gastric cancer, RUNX2 expression levels were analyzed by immunohistochemical staining of 60 cancer tissues and by consulting the Gene Expression Profiling Interactive Analysis (GEPIA) database, which demonstrated the high expression of RUNX2 at both the gene and protein levels in gastric cancer [
33]. In oral squamous cell carcinoma (OSCC), RUNX2 RNA levels were found to be statistically higher in tumor tissues than in normal tissues by qRT-PCR analysis of 40 pathological specimens. A similar result was observed in a comparison between squamous cell carcinoma cells (TCA8113, CAL-27, SCC-9, and TSCCA) and normal oral keratinocytes (NHOK) [
15]. Nickel (Ni) compounds are classified as Group 1 carcinogens, including to the lungs. RUNX2 expression appeared to be increased upon Ni-initiated BEAS-2B transformation, suggesting a potential role in lung tumorigenesis [
60]. RUNX2 expression could also be orchestrated by circular RNA (circRNA)-mediated signaling. In nasopharyngeal carcinoma, circRANBP17 promoted RUNX2 expression by sponging miR-635 [
21]. RUNX2 was overexpressed in tissue samples of bladder urothelial cancer, and immunohistochemistry further demonstrated the positive correlation of high RUNX2 levels with cancer-associated fibroblast biomarkers [
18]. The data of integrating the transcriptomic studies in various cancer types and the matched clinical information were announced and released (University of California, Santa Cruz,
n = 12,839) [
61]. As seen in
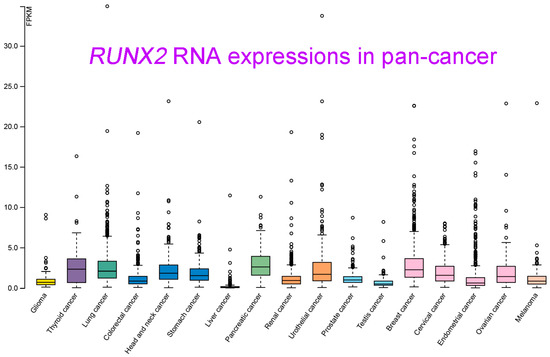
Figure 5,
RUNX2 was shown to be highly upregulated in pancreatic cancer, breast cancer, lung cancer, thyroid cancer, and head and neck cancer. In contrast, lower
RUNX2 levels were detected in liver cancer and testis cancer.
RUNX2 appears to be a prognostic biomarker in many cancer types. In oral cancer patients, a high RUNX2 level was correlated with lymph node metastasis [
57]. Tumor budding has been characterized as a microscopic-finding-based dedifferentiation at the invasive margin in colon cancer. RUNX2 was identified as a constituent of the molecular budding gene signature and contributed to unfavorable relapse-free survival rates in a cohort study of 85 patients with stage II/III disease [
62]. In an exploration of clinical data in colon cancer, RUNX2 was expressed higher in cancer patients with metastasis and shorter survival [
34]. In a clinical study of gastric cancer, patients with positive RUNX2 expression had unfavorable survival, clinical stage, and associated lymph node metastasis [
33]. RUNX2 expression was measured by immunohistochemistry and analyzed for correlations with clinical data in 105 osteosarcoma patients, and it appeared to be an independent predictor of metastasis-free survival and overall survival in a multivariate survival analysis. In addition, RUNX2 and osteopontin expression were strongly correlated at the protein level [
63]. In lung adenocarcinoma, the expression of RUNX2 correlated with a poor hazard ratio, suggesting that RUNX2 plays a clinical role as an independent risk factor for poor survival in lung cancer [
59]. A similar result demonstrated the positive correlation of elevated RUNX2 with poor overall survival of non-small-cell lung cancer patients [
64]. RUNX2 expression was associated with adverse overall survival in a study of 301 renal cell carcinoma patients. In addition, correlations with poor grade and stage were revealed by an analysis of the TCGA database [
12]. In hepatocellular carcinoma, the data from clinicopathological analysis of 89 samples indicated the correlation of RUNX2 expression with metastasis rate and shorter survival period [
23]. An immunohistochemistry-based study of breast cancer tissue samples obtained from 75 patients showed that a high RUNX2 level was significantly associated with poor prognosis, Ki-67 expression, and lymphatic metastasis [
65]. A comprehensive pancancer study integrating cancer patients’ clinical data with RNA expression profiles has been completed and released from the Human Protein Atlas (HPA) [
56,
66,
67,
68,
69] and Kaplan–Meier plotter [
70] databases. The prognostic data of
RUNX2 in different cancer types are listed in
Table 2 (data were adapted with permission from HPA:
https://www.proteinatlas.org/about/licence#citation_guidelines_for_the_human_protein_atlas, accessed on 21 February 2023).
RUNX2 appears to be an inferior prognostic biomarker in cohorts of patients with glioma, colorectal cancer, stomach cancer, pancreatic cancer, renal cancer, urothelial cancer, lung cancer, and cervical cancer. On the other hand, in patients diagnosed with breast and ovarian cancer determined by array, high
RUNX2 expression levels are correlated with better clinical outcomes.