Gastric cancer (GC) is one of the most common and difficult diseases to treat. The study of signaling pathway regulation by microRNA provides information on the mechanisms of GC development and is the basis for biomarker creation. In this study, a circuit of microRNA interactions with signaling pathways was constructed. In most cases, microRNAs in GC regulate the Wnt/b-catenin, PI3K/AKT/mTOR, RAS/RAF/ERK/MAPK, NF-kB, TGF-b, and JAK/STAT pathways. Part of the microRNA acts on several target genes that function in different pathways. This often leads to an intensification of the induced processes. MicroRNAs have also been described that have the opposite effect on different pathways, causing different functional consequences. By acting on several target genes, or genes associated with several pathways, microRNAs can function in a signaling network. The characteristics of microRNAs proposed as candidates for GC biomarkers were analyzed. The currently developed diagnostic and prognostic panels of microRNAs are also considered.
- gastric cancer
- microRNA
- signaling pathways
- markers
1. Introduction
GC is the fourth most morbid and second most fatal cancer in the world and is a serious clinical problem due to late detection, resistance to chemotherapy, and poor prognosis [1]. Currently, new targets for the development of effective approaches to the treatment of GC, as well as informative diagnostic and prognostic biomarkers, are being actively pursued. In this regard, microRNAs are of great interest. MicroRNAs are small non-coding RNAs (18 to 24 nucleotides) that regulate many biological processes, including the development of cancer. MicroRNAs are regulators of signaling pathways, and thus are involved in various processes of a tumor cell, such as proliferation, invasion, migration, and metastasis. By interacting with certain genes in signaling pathways, microRNAs can suppress or induce the development of an oncological process. Thus, the study of the regulation of signaling pathways by using microRNAs provides information on the mechanisms of GC development and is the basis for the creation of diagnostic and prognostic markers. MicroRNAs, in addition to their functional properties, have structural features that ensure their attractiveness as markers—they are weakly subject to degradation in biological fluids, mainly due to their small size and movements within exosomes.
A number of works have been published in which the relationship of microRNAs with metastasis and tumor chemoresistance is considered, and potential GC biomarkers have been presented [2][3][4][5].
The interaction of microRNAs with signaling pathways has a number of features, which consist of different forms of regulation and are associated with the possibility of simultaneous action on several pathways. These features can lead to various functional and clinical consequences that do not follow directly from the action of microRNA on a specific gene. The possibility of microRNA action on several genes, as well as the possibility of genes functioning in several pathways and crosstalk between paths, have not been considered. Accordingly, these features are directly related to the choice of microRNAs for analysis as biomarkers or therapeutic targets. However, these features have not been previously summarized and analyzed. The relevance of a deeper analysis emphasizes the absence of effective and applied molecular systems for diagnostics and prognosis of GC [6]—a role microRNA is potentially capable of performing. It is also important not only to cite, but also to analyze the available data on potential microRNA-based biomarkers, in order to get a clearer picture of the current potential of this technology.
2. Features of the microRNA Interaction with Signaling Pathways in GC
MicroRNAs are regulators of signaling pathways involved in various processes of a tumor cell, such as proliferation, invasion, migration, and colony formation. By interacting with certain genes in signaling pathways, microRNAs can suppress or induce cancer development. This review presents microRNAs involved in the pathogenesis of GC, and their relationship with signaling pathways was traced. Most of the works describe the relationship of microRNA with the Wnt/β-catenin, PI3K/AKT/mTOR, RAS/RAF/ERK/MAPK, NF-κB, TGF-β, and JAK/STAT pathways. These signaling pathways and the microRNAs regulating them are shown in Figure 1.
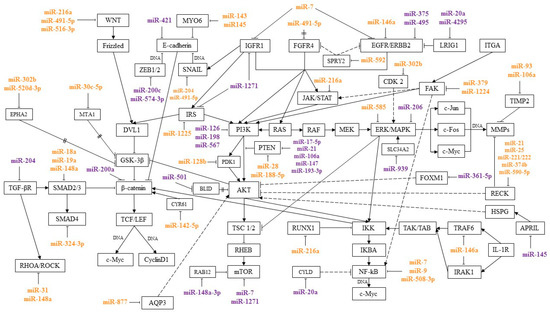
Figure 1. MicroRNAs and signaling pathways regulated by them in GC. miR—associated with metastasis; miR—associated with chemoresistance;
 activation;
activation;  inhibition;
inhibition;  interaction is assumed;
interaction is assumed;  mediated interaction.
mediated interaction.
Activation of the Wnt/β-catenin signaling pathway is found in 10% to 50% of cases and in many GC cell lines. Wnt/β-catenin is one of the main signaling pathways involved in epithelial-mesenchymal transition (EMT) and plays a key role in metastasis. Suppression of the Wnt/β-catenin pathway inhibited the development of metastases in GC models [7][8].
PI3K/AKT/mTOR is involved in the regulation of the cell cycle and processes such as cell growth and metabolism. In a tumor cell, PI3K/AKT/mTOR is one of the main signaling pathways in the regulation of proliferation, invasion, and migration [9]. Protein kinase B (AKT) is one of the central proteins of the PI3K/AKT/mTOR pathway. Overexpression of p-AKT leads to dysregulation of the cell cycle, suppression of apoptosis, and activation of angiogenesis. In GC, increased levels of AKT and p-AKT expression were observed in 74% and 78% of cases. There was also a correlation between the level of p-AKT expression with the depth of tumor invasion and the degree of lymph node involvement [10]. AKT activation leads to phosphorylation of a number of proteins such as GSK3β, BAD, CASP9, Forkhead transcription factors, IκB kinase, and others. Proteins, depending on their type, are activated or inhibited as a result of phosphorylation. Glycogen synthase kinase 3 beta (GSK3β) is a regulator of the Wnt/β-catenin signaling pathway. Phosphorylation inhibits GSK-3, thereby activating the Wnt/β-catenin signaling pathway. AKT is also an activator of the NF-κB signaling pathway. IκB kinase (IKK) is a key component of the NF-κB pathway. Phosphorylation activates IKK and the NF-κB signaling pathway, inducing the expression of genes encoding antiapoptotic proteins [11][12]. In addition, in vitro and in vivo experiments have shown that activation of AKT/IκB/NF-κB induces EMT and leads to the development of metastases [13].
NF-κB is involved in the activation of the migratory and invasive properties of the cell, the regulation of apoptosis, and other cellular processes. In addition, this signaling pathway plays a key role in the regulation of the immune response and the inflammatory response. Activation of the NF-κB pathway not only promotes neoplastic cell transformation and the development of the oncological process, but also allows the tumor to avoid the response of the body’s immune system [14].
The RAS/RAF/ERK/MAPK signaling pathway interacts with PI3K/AKT/mTOR and activates NF-κB. MAPK is one of the key signaling pathways involved in the regulation of proliferative, migratory, and invasive cell properties and angiogenesis [15][16].
It was found that in GC the signaling pathways PI3K/AKT/mTOR, RAS/RAF/ERK/MAPK, and NF-kB can jointly participate in the regulation of a number of cellular processes and suppress the sensitivity of tumor cells to chemotherapy drugs—5-fluorouracil and cisplatin. In experiments on cell lines and tissue samples of GC, it was shown that PI3K/AKT/mTOR activates integrin-linked kinase (ILK), which, in turn, can activate the Ras/c-Raf/MEK1/2/ERK1/2/IκBα pathway and NF-κB. By activating the NF-κB signaling pathway, integrin-linked kinase induces EMT, angiogenesis, cell migration, and invasion, and suppresses apoptosis. ILK overexpression is associated with the depth of tumor invasion and the degree of metastasis to the lymph nodes in GC [17].
There is crosstalk between the NF-κB and JAK/STAT signaling pathways. The transcription factors NF-κB and STAT3 are activated by cytokines and regulate (sometimes jointly) the transcription of genes involved in apoptosis, proliferation, and other cellular processes. In GC, NF-κB and STAT3 act as oncogenes, enhancing the metastatic potential of tumor cells and contributing to the development and progression of the tumor [18].
The Ras homolog family member A/Rho-associated protein kinase 1 (RHOA/ROCK) and TGF-β pathways also induce the invasion and migration of tumor cells, and are associated with lymph node damage and EMT activation [19][20].
Thus, the relationship between the pathways described above forms a signaling network, leading to their joint participation in the regulation of cellular processes. When one pathway is activated, other signaling pathways associated with it can be activated, inducing a cascade of reactions leading, in particular, to suppression of apoptosis, and activation of invasive and migratory properties of a tumor cell.
MicroRNAs, acting on their target genes, can be inhibitors or activators of signaling pathways. For the most part, microRNAs, shown Figure 1, or their target genes interact with key effectors of these pathways, such as PI3K, AKT, PTEN, MAPK, mTOR, WNT, E-cadherin, β-catenin, IKK, and NF-κB.
MicroRNAs that inhibit several target genes, regulate several signaling pathways, and that can be involved in various cellular processes have been described. For example, miR-21 and miR-106 suppress the common target PTEN, as well as genes RECK and TIMP2 that inhibit the metalloproteinases. This activates PI3K/AKT/mTOR and a number of signaling pathways with which metalloproteinases interact.
Some microRNAs can simultaneously be activators of one pathway and blockers of another. Thus, miR-216a blocks the JAK/STAT and Wnt/β-catenin pathways, inhibiting JAK2 and WNT3a, and activates the NF-κB pathway, inhibiting the tumor suppressor gene RUNX1.
In addition, microRNAs regulate signaling pathways by inhibiting components of adjacent signaling cascades. In particular, miR-18a, miR19a, and miR-188-5p have been described as regulators of the Wnt/β-catenin signaling pathway. At the same time, miR-188-5p regulates this pathway, inhibiting PTEN, a key component of the PI3K/AKT/mTOR signaling pathway, and miR-18a and miR19a, inhibiting SMAD2, the TGF-β pathway mediator. Moreover, by inhibiting one target, microRNAs can act on two signaling pathways. These microRNAs include miR-592, which, by suppressing its target SPRY2, activated two signaling pathways, PI3K/AKT/mTOR and RAS/RAF/ERK/MAPK.
A more detailed consideration of the microRNA interaction with their targets and signaling pathways will make it possible to better understand the role of these microRNAs in the regulation of various cellular processes in GC. As mentioned above, a number of microRNAs act on several targets, regulating several signaling pathways. MiR-21 and miR-106a, by inhibiting PTEN, activate the PI3K/AKT/mTOR pathway, which leads to the induction of the proliferative properties of the tumor cell, as well as to the suppression of apoptosis and autophagy. In addition, the targets of miR-21 and miR-106a—RECK and TIMP2—are associated with the Wnt/β-catenin, PI3K/AKT/mTOR, RAS/RAF/ERK/MAPK, NF-κB, JAK/STAT, and Notch pathways. Suppression of RECK and TIMP2 leads to activation of metalloproteinases and destruction of the extracellular matrix and basement membrane of cells, which are predictors of the metastatic process [21][22][23][24][25][26][27]. These microRNAs, by inhibiting various targets, affect a number of signaling pathways, which leads to the suppression of apoptosis and promotes the development of metastases.
MiR-302b is a suppressor of tumor growth and metastasis in GC. A decreased level of miR-302b expression is associated with the involvement of regional lymph nodes in the metastatic process, peritoneal carcinomatosis, and the development of distant metastases. Direct targets of miR-302b include CDK2 and EPHA2. Transfection with miR-302b analogs inhibited the RAS/RAF/ERK/MAPK and Wnt/β-catenin signaling pathways in GC cells [28][29][30].
MiR-20a is a direct inhibitor of LRIG1. Suppression of LRIG1 leads to the activation of EGFR and signaling pathways PI3K/AKT/mTOR and RAS/RAF/ERK/MAPK. This activates the ATP-binding cassette transporter P-gp (ABCB1), which causes an increase in the outflow of drugs from the cell. In addition, the anti-apoptotic protein BCL2 is also induced. The CYLD gene is another target of miR-20a. Suppression of CYLD activates the NF-κB signaling pathway and anti-apoptotic proteins Livin and Survivin.
Due to the action of these mechanisms, miR-20a overexpression caused an increase in the efflux of chemotherapy drugs from cultured GC cells, as well as apoptosis suppression, which led to the development of tumor cell chemoresistance [31][32].
Decreased miR-20a expression was shown to inhibit Wnt/β-catenin and RAS/RAF/ERK/MAPK signaling pathways. This, in turn, led to inhibition of growth, as well as the invasive and migratory properties of GC cells by in vitro experiments [33].
In experiments on GC cell lines, miR-491-5p suppressed cell migration and proliferation and promoted apoptosis. MiR-491-5p was originally described as an inhibitor of the antiapoptotic factor BCL-XL. MiR-491-5p has been shown to inhibit ERK1/2 and AKT. Later, it was found that direct targets of miR-491-5p are SNAIL and WNT3A. Overexpression of miR-491-5p inhibited FGFR4, N-cadherin, fibronectin (FN1), c-Myc, TCF-1, and CyclinD and activated E-cadherin, as a result of which miR-491-5p inhibited EMT [34][35]. Thus, miR-491-5p inhibits the signaling pathways PI3K/AKT/mTOR, Wnt/β-catenin, and RAS/RAF/ERK/MAPK, which leads to suppression of metastases development and induces apoptosis.
MiR-7 is a direct inhibitor of a number of targets—mTOR, EGFR, IGF1R, and RELA—and thus blocks several signaling pathways. By inhibiting its target mTOR (PI3K/AKT/mTOR pathway), miR-7 induces apoptosis and suppresses tumor growth in in vivo experiments [36]. As a result of suppression of its target RELA, miR-7 inhibits the NF-κB signaling pathway and genes associated with metastasis: VNT, ICAM-1, VCAM-1, MMP-2, MMP-9, and VEGF [37][38].
The genes IGF1R, IRS1, mTOR, and BCL2 have been identified as direct targets of miR-1271 in GC [39]. IGF1R and IRS1 belong to the IGF/IGF1R/IRS1 signaling pathway, which is often considered as part of the PI3K/AKT/mTOR pathway and is involved in the regulation of cell proliferation and apoptosis [40]. IGF/IGF1R/IRS1 is an activator of the PI3K/AKT/mTOR pathway. As a result of PI3K/AKT/mTOR activation, the antiapoptotic protein BCL2 is induced [41]. Thus, by acting on a number of targets, miR-1271 inhibits cell proliferation and induces apoptosis [39].
Despite the fact that both miR-7 and miR-1271 are inhibitors of the same signaling pathway (IGF/IGF1R/IRS1), they act as regulators of different cellular processes: miR-7 is described as an EMT inhibitor, and miR-1271 as an inducer apoptosis. Thus, even if microRNAs initially act on the same signaling pathway, they can regulate different cellular processes.
Some microRNAs are capable of multidirectional effects on signaling pathways. In particular, miR-216a and miR-148a have been described as inhibitors of one signaling pathway and activators of another. MiR-216a inhibits the Wnt/β-catenin and JAK/STAT signaling pathways by suppressing the direct targets Wnt3a and JAK2. In Tao et al., overexpression of miR-216a led to the activation of E-cadherin and inhibited EMT in in vitro experiments. The decreased level of miR-216a expression correlated with metastasis to the lymph nodes in patients with GC [42][43]. At the same time, miR-216a is an inducer of the NF-κB signaling pathway. Overexpression of miR-216a activated the NF-κB signaling pathway by suppressing RUNX1 and led to an increase in the expression levels of CyclinD1, BCL2, MMP-2, and MMP-9, inducing the migratory, proliferative, and invasive properties of cells [44]. Therefore, miR-216a plays a dual role in the progression of GC. On the one hand, this microRNA inhibits the Wnt/β-catenin and JAK/STAT signaling pathways, acting as a tumor suppressor and preventing metastasis; on the other hand, it promotes the development of metastases by activating the NF-κB pathway.
MiR-148a inhibits the TGF-β and RHOA/ROCK pathways, suppressing the targets of SMAD2 and ROCK1. The TGF-β signaling pathway is one of the regulators of EMT. TGF-β activation correlates with an increase in the metastatic potential of GC cells. MiR-148a, by inhibiting SMAD2 and the TGF-β pathway, led to the activation of E-cadherin and suppression of WNT. This caused suppression of EMT and reduced the ability of GC cells to invade and migrate [45]. In addition, in experiments on GC cells, miR-148a inhibited ROCK1-induced invasion and migration of GC cells [46]. At the same time, miR-148a activates m-TORC1 (PI3K/AKT/mTOR pathway), inhibiting RAB12. The direct target of miR-148a, RAB12, is a potent autophagy inducer. Autophagy is the degradation of cell organelles and proteins used as a source of additional energy. This allows the tumor cell to survive in adverse conditions. In in vitro experiments, miR-148a inhibited RAB12 and, as a consequence, autophagy. As a result, overexpression of miR-148a increased the sensitivity of tumor cells to cisplatin [47].
Therefore, by exerting an opposite effect on different pathways—inhibiting the TGF-β and RHOA/ROCK pathways, and activating PI3K/AKT/mTOR—miR-148a causes various functional consequences: It suppresses the development of metastases and increases the sensitivity of tumor cells to chemotherapy.
As noted above, a number of microRNAs regulate signaling pathways by interacting with mediators of adjacent signaling cascades. Thus, miR-188-5p inhibits PTEN, while AKT is activated. The AKT, in addition to the PI3K/AKT/mTOR pathway, activates the Wnt/β-catenin pathway, suppressing GSK3β. In cancer cell lines, overexpression of miR-188-5p inhibited PTEN and GSK3β, resulting in an increase in the expression levels of gene-effectors of the Wnt/β-catenin signaling pathway. MiR-188-5p enhanced the cells’ ability to migrate and invade and their metastatic potential [48].
MiR-18a, miR19a, and miR-324-3p act on components of the TGF-β pathway and activate the Wnt/β-catenin pathway. SMAD2, a direct target of miR-18a and miR19a, is associated with the TGF-β pathway. Moreover, SMAD2 is an inhibitor of β-catenin (Wnt/β-catenin pathway). It was experimentally shown that miR-18a and miR-19b, by inhibiting SMAD2, increased the expression levels of β-catenin, C-Myc, and axin2, activated the Wnt/β-catenin signaling pathway and the metastatic potential of GC cells. Induced expression of miR-18a/19a significantly enhanced the ability of GC cells to migrate and proliferate [49]. The direct target of miR-324-3p is SMAD4. In in vitro experiments, miR-324-3p inhibited apoptosis and promoted proliferation, migration, and survival of GC cells [50].
The APRIL gene has been identified as a direct target of miR-145 in GC. In experiments on cell lines, it was shown that APRIL, interacting with Heparan Sulfate Proteoglycan (HSPG), phosphorylates AKT. This, in turn, activates the NF-κB signaling pathway and the effectors of this pathway—BCL2 and BCL-XL (inhibitors of apoptosis). Suppression of APRIL led to inactivation of AKT and the NF-κB signaling pathway, and promoted apoptosis [51].
By suppressing its direct target PDK1, miR-128b also affected AKT, while inhibiting the NF-κB pathway in GC. It was shown that miR-128b inhibited p-AKT and NF-κB in GC cells and inactivated the PDK1/AKT/NF-κB axis [52].
MiR-361-5p is a FOXM1 inhibitor that affects the PI3K/AKT/mTOR pathway. The FOXM1 transcription factor belongs to the Forkhead family. According to Tian et al., FOXM1 inhibits the PI3K/AKT/mTOR pathway. In vitro experiments miR-361-5p inhibited FOXM1 and led to an increase of p-AKT and mTOR. As a result of activation of the PI3K/AKT/mTOR signaling pathway, miR-361-5p inhibited autophagy and increased the sensitivity of tumor cells to chemotherapy [53].
The direct target of miR-582, FOXO3, also belongs to the Forkhead family. As a result of FOXO3 suppression, miR-582 induced the PI3K/AKT/mTOR signaling pathway. This, in turn, led to the activation of the transcription factor SNAIL, which is an inhibitor of E-cadherin and is associated with the Wnt/β-catenin pathway [54]. Thus, by activating the PI3K/AKT/SNAIL signaling pathway, miR-582 led to the induction of EMT, which promoted the growth, invasion, and GC metastasis [55].
The direct target of miR-877, aquaporin 3 (AQP3), is also a regulator of the PI3K/AKT/mTOR pathway. By activating the PI3K/AKT/SNAIL axis, AQP3 induces EMT. In addition, AQP3 has been shown to enhance the resistance of GC cells to cisplatin. In vitro experiments, overexpression of miR-877 inhibited AQP3, resulting in suppression of EMT, invasion and proliferation of GC cells, and also induced apoptosis [56].
Thus, by inhibiting genes for some signaling pathways, miR-18a, miR-19a, miR-128b, miR-145, miR-188-5p, miR-324, miR-361-5p, miR-582, and miR-877 acted on other cascades connected with these paths.
By inhibiting one target, microRNA regulated two signaling pathways. In particular, it was shown that miR-592, as a result of suppression of its target SPRY2, activated two signaling pathways, PI3K/AKT/mTOR and RAS/RAF/ERK/MAPK. This led to the induction of the proliferative, invasive, and migratory properties of GC cells [57]. MiR-1224, being a direct inhibitor of focal adhesion kinase (FAK), inhibits the activity of the JAK/STAT and NF-κB signaling pathways and inhibits EMT. In addition, in experiments on GC cell lines, a decrease in miR-1224 expression led to an increase in WNT and ZEB1 expression levels, a decrease in E-cadherin levels, and induced cell migration. It was shown in vivo that miR-1224 prevents the development of distant metastases [58]. By influencing different genes and pathways, microRNAs may be able to exert a unidirectional effect on cellular processes.
Thus, by acting on several target genes or genes associated with several pathways, microRNAs can function in the format of a signaling network. It is still impossible to say with certainty on how many pathways a particular microRNA affects and to what extent. However, the signaling pathways in which the greatest amount of microRNA is involved in regulation during GC are indicated. In most cases, according to the available data, microRNAs regulate the Wnt/β-catenin, PI3K/AKT/mTOR, RAS/RAF/EKK/MAPK, NF-kB, TGF-β, and JAK/STAT pathways. As follows from the data in Figure 1, a significant proportion of microRNAs associated with metastasis regulate the Wnt/β-catenin pathway. The largest amount of microRNAs associated with GC chemoresistance is involved in the regulation of the PI3K/AKT/mTOR pathway.
3. MicroRNA as GC Prognostic and Diagnostic Markers
MicroRNAs can act as convenient clinical biomarkers, given their high stability in different body environments and differential expression in GC. Circulating and tissue microRNAs can allow detection of GC at an early stage, assessment of the course of the disease in dynamics, including determining the likelihood of relapse and (or) metastasis, and prediction of the tumor response to chemotherapy. Thus, it is expected that the use of microRNAs as markers will increase the sensitivity and specificity of diagnostic and prognostic tests for GC [59].
Currently, numerous studies are being carried out, including microarrays, to identify differentially expressed microRNAs in GC. In many articles where the relationship of microRNAs with signaling pathways was considered, the authors report the possibility of using various microRNAs as prognostic or diagnostic markers. In these studies, it was shown that the expression level of the investigated microRNAs significantly differed in the GC tissue, as compared to the unaffected mucosa, and was associated with metastasis. For example, Guan et al. found that miR-93 overexpression was correlated with lymph node metastasis in GC (p < 0.01) [60]. Cha et al. reported that decreased miR-140-5p expression correlates with lymph node metastasis (p = 0.018) and is further associated with a poor prognosis (progression free survival (PFS) and overall survival (OS), p < 0.05) [61].
Table 1 shows the microRNAs characteristics considered as candidates of GC biomarkers. In addition to microRNAs with known interactions with signaling pathways, this section also includes microRNAs for which such a relationship was not considered.
| MicroRNA | Characteristics of microRNAs as Candidates for GC Markers | Material/Method | Reference |
|---|---|---|---|
| miR-9 | Lymph nodes metastasis (p < 0.001) and distant metastases (p = 0.022). |
GC tissue samples | [62] |
| miR-19b miR-106a | Diagnostics: AUC = 0.814, sensitivity = 95%, and specificity = 90%. |
Circulating exosomal microRNAs | [63] |
| miR-21 miR-106a |
Difference in expression in gastric and non-cancerous cancers (p < 0.001). |
Gastric juice | [64] |
| miR-23a miR-135 |
Diagnostics: miR-23a specificity, sensitivity, and AUC are 67.95, 87.50, and 0.805%, respectively, for miR-135 at 73.08, 82.50, and 0.824%, respectively. |
Serum | [65] |
| miR-24 miR-101 |
OS (p < 0.01). | GC tissue samples | [66] |
| miR-29c | Decreased expression level in tumor tissue compared to unaffected mucosa (p < 0.0001). | GC tissue samples | [67] |
| miR-30c-5p | Lymph node metastasis (p = 0.014). | GC tissue samples | [68] |
|
miR-34a miR-146a |
For miR-34a lymph nodes and distant metastasis (p = 0.001) For miR-146a lymph nodes and distant metastasis (p = 0.002). |
GC tissue samples |
[69] |
| miR-93 | Lymph node metastasis (p < 0.01). | GC tissue samples | [60] |
| miR-101-3p | When distinguishing atrophic gastritis and gastric cancer: AUC = 0.8749, sensitivity = 72.09%, and specificity = 86.49%. |
Serum | [70] |
| miR-106 | Sensitivity = 0.71, specificity = 0.82, and AUC = 0.80. | Serum, tissue, plasma and gastric juice | [71] |
| miR-106a | Lymph node metastasis (p = 0.002), vascular invasion (p = 0.017), and the depth of tumor invasion (p = 0.009). |
GC tissue samples | [27] |
| miR-107 | The expression level was increased in adenoma with high-grade dysplasia p = 0.006 and in the early stages of GC p = 0.03. | Microarrays, validation by RT-PCR | [72] |
| miR-129 | Significantly lower levels of miR-129-1-3p (p = 0.007), miR-129-2-3p (p = 0.003), and a combination of two microRNAs (p = 0.003) in GC patients compared to patients with benign stomach diseases. |
In gastric juice | [72] |
| miR-140-5p | Lymph node metastasis (p = 0.018); survival—PFS, p < 0.05, OS, p < 0.05). |
GC tissue samples | [61] |
| miR-155 miR-223 |
Atrophic gastritis (p < 0.0001); gastric cancer (p < 0.05). | GC and AG tissue samples | [73] |
| miR-181a | Lymph node metastasis (p = 0.0124); distant metastases (p = 0.0376). |
GC tissue samples | [74] |
| miR-181d | Lymph node metastasis (p < 0.05) and overall survival (p = 0.001). |
GC tissue samples | [75] |
| miR-196a | Survival: (p = 0.032, HR = 3.057, 95% CI = 1.1–8.495); Diagnostics: AUC = 0.864, sensitivity 69.5%, specificity 97.6%; metastasis p < 0.001. | Plasma | [76] |
| miR-196b | Survival: p = 0.042, HR = 2.914, 95% CI = 1.036–8.174; diagnostics: AUC = 0.811, sensitivity 62.2%, and specificity 96.1%. |
Plasma | [76] |
| miR-197 | Depth of invasion (p = 0.005), lymph node metastases (p = 0.004). |
GC tissue samples | [77] |
| miR-200c | Had a prognostic value and moderately diagnostic. Expression data were inconsistent. | Meta-analysis | [78] |
| miR-302b | Lymph node metastases (p = 0.003), OS (HR = 1.86; 95% CI, 1.11–3.14; p = 0.021). |
GC tissue samples | [29] |
| miR-330-3p | Lymph node metastasis (p < 0.001). | Serum, GC tissue samples | [79] |
| miR-361-5p | Lymph node metastasis, distant metastases development (p < 0.001). |
GC tissue samples | [80] |
| miR-376a | Metastasis to regional lymph nodes (p = 0.02) and poor prognosis (p = 0.02). |
Tissues and cell lines of GC | [81] |
| miR-379 | Lymph node metastasis (p < 0.001), OS (p = 0.0007), and PHS (p = 0.0002). |
GC tissue samples | [82] |
| miR-421 | Difference in expression among patients with benign and malignant gastric diseases (p < 0.001). | Gastric juice | [64] |
| OS (p = 0.016, HR 2.586, 95% CI 1.194–5.599) and RFS (p = 0.014, HR 2.465, 95% CI 1.201–5.060). |
GC tissue samples | [83] | |
| Early stages. Sensitivity 96.67; specificity 95.56; AUC 0.981 (0.942–0.997); p <0.0001. |
Serum | [84] | |
| miR-484 | Lymph node metastasis (p = 0.015), distant metastases development (p = 0.005), stage of the disease (p = 0.002), and degree of differentiation (p = 0.006). |
GC tissue samples | [85] |
| miR-519a | Lymph node metastasis, degree of differentiation, and stage of the disease (p < 0.05); OS (p = 0.002). |
GC tissue samples | [86] |
| miR-520a-3p | Depth of tumor invasion (p < 0.001) and stage of the disease (p < 0.05). |
GC tissue samples | [87] |
| miR-552 | Lymph node metastasis (p = 0.018) and OS (p = 0.011), HR = 5.657, 95% CI 1.619–19.761. |
GC tissue samples | [88] |
| miR-585 | Depth of tumor invasion (p < 0.010), lymph node metastasis (p < 0.002). |
GC tissue samples | [89] |
| miR-601 | Invasion, lymph node metastasis, and the distant metastases development (p < 0.05); OS (p = 0.001). |
GC tissue samples | [90] |
| miR-1225-5p | Depth of tumor invasion (p = 0.016), spread of metastases to lymph nodes (p = 0.002), and development of distant metastases (p = 0.01). | GC tissue samples | [91] |
| miR-1236-3p | Lymph node metastasis (p = 0.005), disease stage (p = 0.001), and degree of differentiation (p = 0.001). | GC tissue samples | [92] |
| Potential markers of response to chemotherapy | |||
| miR-27b miR-508-5p |
Response to chemotherapy (p = 0.02 and p = 0.04, respectively). | GC tissue samples | [93] |
| miR-939 | Potential marker of sensitivity to chemotherapy; AUC = 0.777, p < 0.001. | GC tissue samples | [94] |
| MicroRNA panels | |||
| miR-7-2 miR-9-3 miR-548o miR-1255a miR-3687 |
Patient survival AUC = 0.9 (HR, 2.840; 95% CI, 1.937–4.162; p < 0.01). |
TCGA database | [95] |
| miR-143-3p miR-146a miR-451a miR-501-3p | Predicting the development of lymph node metastases in GC; AUC = 0.822 (95% CI, 0.758 to 0.875), specificity = 87.78%, and sensitivity = 63.33%. | Serum | [96] |
| miR-22-5p miR-132-3p miR-200a-3p miR-485-3p miR-2965p | Suggested for diagnostics of GC AUC= 0.724. | Serum | [97] |
| miR-10b-5p miR-20a-3p miR-132-3p miR-185-5p miR-195-5p miR-296-5p | Suggested for diagnostics of GC AUC = 0.702. | Serum | [98] |
AUC—area under the curve; PFS—progression-free survival; RFS—relapse-free survival; OS—overall survival; HR—hazard ratio; CI—confidence interval; RR—relative risk.
The microRNAs listed in Table 1 have different levels of evidence for their acceptability as markers. The closest to practical use are marker panels. However, data on a number of individual microRNAs demonstrate their significant potential as markers. These microRNAs, presented in Table 1, include miR-101-3p, miR-106a, miR-135, miR-140-5p, miR-196, and miR-552, which have a significant relationship with the clinical characteristics under study (given in Table 1). MiR-421, when associated with a number of clinical features, is most significantly associated with the early stage of GC, which indicates its promising potential as a diagnostic marker. It is also worth highlighting miR-129, the expression of which in GC differs from its expression among patients with benign gastric diseases.
At the same time, not all microRNAs positioned as candidates for GC markers have been studied from the point of view of possible expression changes in other gastric diseases. In particular, microRNA regulation can be impaired not only in malignant tumors but also under Helicobacter pylori infection, chronic gastritis, atrophic gastritis, intestinal metaplasia, and early dysplasia [99]. For example, according to Link et al., expression levels of miR-155 and miR-223 were increased in both GC and atrophic gastritis. No statistically significant differences in the expression levels of miR-155 and miR-223 were found between GC samples and atrophic gastritis [73]. Some microRNAs associated with GC, for example, miR-16, miR-17-5p, miR-20a, miR-22, miR-126, miR-132, miR-143, miR-191, miR-195, and miR-200, are also considered as markers of other (including frequent) diseases, such as cerebral atherosclerosis, endometriosis, and heart disease [100][101][102][103].
In addition, the results of published studies on some microRNAs are inconsistent. MiR-216a, according to Tao et al. [42], suppresses the metastases development. On the other hand, Wu et al. report that miR-216a promoted the metastases development and was associated with a poor prognosis [44]. MiR-135b, according to different authors, is considered both an apoptosis inhibitor and an inducer [104][105]. As shown by the results of the meta-analysis, the value of miR-200c as a diagnostic and prognostic marker cannot be considered definitively determined [78].
The circulating microRNAs constitute a significant part of microRNAs investigated as potential markers. They have generated significant interest, since their definition refers to minimally invasive diagnostic methods. Many of the circulating microRNAs have high sensitivity, specificity AUC, RR, etc. (Table 1). However, there are issues with circulating microRNA that require clarification. In particular, when comparing the expression levels of miR-196a and miR-196b in the blood serum and tumor tissues of the stomach, the correlation coefficient was 0.53 and 0.45. That is, the expression level of circulating microRNA in a significant number of cases did not correspond to the expression levels of tissue miR-196a and miR-196b [76].
In order to increase the sensitivity and specificity of microRNAs as markers, panels are being developed that include several microRNAs. One of the earliest panels was proposed back in 2011. Liu et al. developed a panel of five serum microRNAs (miR-1, miR-20a, miR-27a, miR-34, and miR-423-5p) for GC detection. For this panel, AUC = 0.879. This was higher than for other biomarkers, including CEA (AUC = 0.503) and CA19-9 (AUC = 0.6) [106]. A panel of five serum microRNAs (miR-16, miR-25, miR-92a, miR-451, and miR-486-5p) was proposed by Zhu et al. The expression level of these microRNAs was increased in GC patients compared to the control group. ROC (receiver operating characteristic) analysis showed high diagnostic accuracy for the early stage of GC [107]. Jiang et al. examined a panel of four microRNAs (miR-143-3p, miR-146a, miR-451a, and miR-501-3p) as non-invasive biomarkers to predict the lymph node metastases development in GC. The experimental group included 279 people; the validation group included 180 people. The characteristics of this panel in the validation group were AUC = 0.822 (95% CI, 0.758 to 0.875), specificity = 87.78%, and sensitivity = 63.33%. Moreover, Kaplan–Meier analysis showed that patients with lymph-node metastases and low levels of miR-146a and miR-451a expression had the worst OS (p < 0.05) [96].
Expression databases are often used in panel design. Zhao et al. used clinical and microRNA-seq data from patients with gastric adenocarcinoma (n = 310) downloaded from The Cancer Genome Atlas (TCGA) database. A microRNA prognostic panel (hsa-mir-7-2, hsa-mir-9-3, hsa-mir-548o, hsa-mir-1255a, and hsa-mir-3687) was developed for GC patients, but panel validation by RT-PCR was not performed. For validation, a verification group was formed using patient data from the TCGA database. In the experimental group, the AUC was 0.939, in the verification—0.901. The sensitivity and specificity of the panel were not indicated in the paper text. From the presented graphs of the ROC analysis, the sensitivity was about 90% for both groups, the specificity is about 75%. The Kaplan–Meier log rank test showed that low-risk patients had significantly longer survival times than high-risk patients (HR, 2.840; 95% CI, 1.937–4.162; p < 0.01). An appropriate formula was developed to use this panel. However, the authors have shown that a history of relapse and age over 65 years are independent prognostic factors [95].
In 6% to 16% of cases, gastric adenocarcinoma is accompanied by infection with the Epstein–Barr virus (EBV), which expresses microRNAs in the tumor [108]. Treece et al. developed the GastroGenus miR panel, which includes microRNAs encoded by the Epstein–Barr virus (EBV) and cancer-specific microRNAs. In GC tissues infected with EBV, an increased expression of EBV-encoded microRNAs (p < 0.006) was compared with uninfected ones. Concomitant dysregulation of four hsa-miRs expression (p < 0.00125) was observed with overexpression of EBV-microRNA. There was formed a hypothesis that EBV infection could affect the regulation of GC cell signaling pathways [109]. It is assumed that this panel will differentiate EBV-positive GC from EBV-negative one.
The number of microRNAs studied in connection with the clinical manifestations of GC is steadily increasing. Diagnostic and prognostic microRNAs and their panels are being actively developed. However, to date, no microRNA (microRNA panel) has been found that could be used in clinical practice.
In the European Society for Medical Oncology (ESMO) version, as diagnostic and prognostic criteria of GC recommended indicators of histology, instrumental studies, and her2-status of the tumor [6]. This circumstance underlines the relevance of further studies of microRNAs as candidates for such biomarkers. Some of the microRNAs presented in this work can be selected for verification or combined into new diagnostic or prognostic panels.
This entry is adapted from the peer-reviewed paper 10.3390/diagnostics10110891
References
- Bhat, S.A.; Majid, S.; Rehman, M.U. Scenario and future prospects of microRNAs in gastric cancer: A review. Iran. J. Basic Med. Sci. 2019.
- Kim, S.; Bae, W.J.; Ahn, J.M.; Heo, J.H.; Kim, K.M.; Choi, K.W.; Sung, C.O.; Lee, D. MicroRNA signatures associated with lymph node metastasis in intramucosal gastric cancer. Mod. Pathol. 2020.
- Zhao, X.; Hu, G.F.; Shi, Y.F.; Xu, W. Research progress in microRNA-based therapy for gastric cancer. OncoTargets Ther. 2019, 12, 11393–11411.
- Chen, C.; Tang, X.; Liu, Y.; Zhu, J.; Liu, J. Induction/reversal of drug resistance in gastric cancer by non-coding RNAs (Review). Int. J. Oncol. 2019, 54, 1511–1524.
- Yuan, H.L.; Wang, T.; Zhang, K.H. MicroRNAs as potential biomarkers for diagnosis, therapy and prognosis of gastric cancer. OncoTargets Ther. 2018, 11, 3891.
- ESMO Guidelines Committee. Gastric Cancer Treatment Recommendations. Available online: https://www.esmo.org/guidelines/gastrointestinal-cancers/gastric-cancer/eupdate-gastric-cancer-treatment-recommendations2 (accessed on 8 October 2020).
- Jackstadt, R.; Hodder, M.C.; Sansom, O.J. WNT and β-Catenin in Cancer: Genes and Therapy. Annu. Rev. Cancer Biol. 2020, 4, 177–196.
- Weng, J.; Li, S.; Lin, H.; Mei, H.; Liu, Y.; Xiao, C.; Zhu, Z.; Cai, W.; Ding, X.; Mi, Y.; et al. PCDHGA9 represses epithelial-mesenchymal transition and metastatic potential in gastric cancer cells by reducing β-catenin transcriptional activity. Cell Death Dis. 2020, 11, 1–18.
- Jiang, N.; Dai, Q.; Su, X.; Fu, J.; Feng, X.; Peng, J. Role of PI3K/AKT Pathway in Cancer: The Framework of Malignant Behavior; Springer: Amsterdam, The Netherlands, 2020; Volume 47, ISBN 0123456789.
- Matsuoka, T.; Yashiro, M. The role of PI3K/Akt/mTOR signaling in gastric carcinoma. Cancers 2014, 6, 1441–1463.
- Fresno Vara, J.Á.; Casado, E.; de Castro, J.; Cejas, P.; Belda-Iniesta, C.; González-Barón, M. P13K/Akt signalling pathway and cancer. Cancer Treat. Rev. 2004, 30, 193–204.
- Chen, H.T.; Liu, H.; Mao, M.J.; Tan, Y.; Mo, X.Q.; Meng, X.J.; Cao, M.T.; Zhong, C.Y.; Liu, Y.; Shan, H.; et al. Crosstalk between autophagy and epithelial-mesenchymal transition and its application in cancer therapy. Mol. Cancer 2019, 18, 1–19.
- Li, J.; Deng, Z.; Wang, Z.; Wang, D.; Zhang, L.; Su, Q.; Lai, Y.; Li, B.; Luo, Z.; Chen, X.; et al. Zipper-interacting protein kinase promotes epithelial-mesenchymal transition, invasion and metastasis through AKT and NF-κB signaling and is associated with metastasis and poor prognosis in gastric cancer patients. Oncotarget 2015.
- Taniguchi, K.; Karin, M. NF-B, inflammation, immunity and cancer: Coming of age. Nat. Rev. Immunol. 2018, 18, 309–324.
- Cao, Z.; Liao, Q.; Su, M.; Huang, K.; Jin, J.; Cao, D. AKT and ERK dual inhibitors: The way forward? Cancer Lett. 2019, 459, 30–40.
- van de Stolpe, A. Quantitative measurement of functional activity of the pi3k signaling pathway in cancer. Cancers 2019, 11, 293.
- Tseng, P.C.; Chen, C.L.; Shan, Y.S.; Chang, W.T.; Liu, H.S.; Hong, T.M.; Hsieh, C.Y.; Lin, S.H.; Lin, C.F. An increase in integrin-linked kinase noncanonically confers NF-κB-mediated growth advantages to gastric cancer cells by activating ERK1/2. Cell Commun. Signal. 2014.
- Sokolova, O.; Naumann, M. NF-κB signaling in gastric cancer. Toxins 2017, 9, 119.
- Nam, S.; Kim, J.H.; Lee, D.H. RHoA in gastric cancer: Functional roles and therapeutic potential. Front. Genet. 2019, 10, 438.
- Hao, Y.; Baker, D.; Dijke, P. Ten TGF-β-mediated epithelial-mesenchymal transition and cancer metastasis. Int. J. Mol. Sci. 2019, 20, 2767.
- Zhou, H.; Liu, H.; Jiang, M.; Zhang, S.; Chen, J.; Fan, X. Targeting MicroRNA-21 Suppresses Gastric Cancer Cell Proliferation and Migration via PTEN/Akt Signaling Axis. Cell Transplant. 2019, 28, 306–317.
- Gu, Y.; Fei, Z.; Zhu, R. MiR-21 modulates cisplatin resistance of gastric cancer cells by inhibiting autophagy via the PI3K/Akt/mTOR pathway. Anticancer Drugs 2020.
- Fang, Y.; Shen, H.; Li, H.; Cao, Y.; Qin, R.; Long, L.; Zhu, X.; Xie, C.; Xu, W. MiR-106a confers cisplatin resistance by regulating PTEN/Akt pathway in gastric cancer cells. Acta Biochim. Biophys. Sin. 2013.
- Hong, K.J.; Wu, D.C.; Cheng, K.H.; Chen, L.T.; Hung, W.C. RECK Inhibits Stemness Gene Expression and Tumorigenicity of Gastric Cancer Cells by Suppressing ADAM-Mediated Notch1 Activation. J. Cell. Physiol. 2014.
- Zhang, Z.; Li, Z.; Gao, C.; Chen, P.; Chen, J.; Liu, W.; Xiao, S.; Lu, H. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab. Investig. 2008.
- Bautista-Sánchez, D.; Arriaga-Canon, C.; Pedroza-Torres, A.; De La Rosa-Velázquez, I.A.; González-Barrios, R.; Contreras-Espinosa, L.; Montiel-Manríquez, R.; Castro-Hernández, C.; Fragoso-Ontiveros, V.; Álvarez-Gómez, R.M.; et al. The Promising Role of miR-21 as a Cancer Biomarker and Its Importance in RNA-Based Therapeutics. Mol. Ther.-Nucleic Acids 2020, 20, 409–420.
- Zhu, M.; Zhang, N.; He, S.; Lui, Y.; Lu, G.; Zhao, L. MicroRNA-106a targets TIMP2 to regulate invasion and metastasis of gastric cancer. FEBS Lett. 2014.
- Liu, F.Y.; Wang, L.P.; Wang, Q.; Han, P.; Zhuang, W.P.; Li, M.J.; Yuan, H. miR-302b regulates cell cycles by targeting CDK2 via ERK signaling pathway in gastric cancer. Cancer Med. 2016.
- Tang, L.; Hu, H.; He, Y.; Mcleod, H.L.; Xiao, D.; Chen, P.; Shen, L.; Zeng, S.; Yin, X.; Ge, J.; et al. The relationship between miR-302b and EphA2 and their clinical significance in gastric cancer. J. Cancer 2018, 9, 3109–3116.
- Huang, J.; He, Y.; Mcleod, H.L.; Xie, Y.; Xiao, D.; Hu, H.; Chen, P.; Shen, L.; Zeng, S.; Yin, X.; et al. miR-302b inhibits tumorigenesis by targeting EphA2 via Wnt/ β-catenin/EMT signaling cascade in gastric cancer. BMC Cancer 2017.
- Zhou, L.; Li, X.; Zhou, F.; Jin, Z.; Chen, D.; Wang, P.; Zhang, S.; Zhuge, Y.; Shang, Y.; Zou, X. Downregulation of leucine-rich repeats and immunoglobulin-like domains 1 by microRNA-20a modulates gastric cancer multidrug resistance. Cancer Sci. 2018.
- Zhu, M.; Zhou, X.; Du, Y.; Huang, Z.; Zhu, J.; Xu, J.; Cheng, G.; Shu, Y.; Liu, P.; Zhu, W.; et al. MiR-20a induces cisplatin resistance of a human gastric cancer cell line via targeting CYLD. Mol. Med. Rep. 2016.
- Sun, H.; Wang, Q.; Yuan, G.; Quan, J.; Dong, D.; Lun, Y.; Sun, B. Hsa_circ_0001649 restrains gastric carcinoma growth and metastasis by downregulation of miR-20a. J. Clin. Lab. Anal. 2020.
- Yu, T.; Wang, L.N.; Li, W.; Zuo, Q.F.; Li, M.M.; Zou, Q.M.; Xiao, B. Downregulation of miR-491-5p promotes gastric cancer metastasis by regulating SNAIL and FGFR4. Cancer Sci. 2018.
- Sun, R.; Liu, Z.; Tong, D.; Yang, Y.; Guo, B.; Wang, X.; Zhao, L.; Huang, C. MiR-491-5p, mediated by Foxi1, functions as a tumor suppressor by targeting Wnt3a/β-catenin signaling in the development of gastric cancer. Cell Death Dis. 2017.
- Xu, N.; Lian, Y.J.; Dai, X.; Wang, Y.J. miR-7 Increases Cisplatin Sensitivity of Gastric Cancer Cells Through Suppressing mTOR. Technol. Cancer Res. Treat. 2017.
- Ye, T.; Yang, M.; Huang, D.; Wang, X.; Xue, B.; Tian, N.; Xu, X.; Bao, L.; Hu, H.; Lv, T.; et al. MicroRNA-7 as a potential therapeutic target for aberrant NF-κB-driven distant metastasis of gastric cancer. J. Exp. Clin. Cancer Res. 2019.
- Zhao, X.; Dou, W.; He, L.; Liang, S.; Tie, J.; Liu, C.; Li, T.; Lu, Y.; Mo, P.; Shi, Y.; et al. MicroRNA-7 functions as an anti-metastatic microRNA in gastric cancer by targeting insulin-like growth factor-1 receptor. Oncogene 2013.
- Yang, M.; Shan, X.; Zhou, X.; Qiu, T.; Zhu, W.; Ding, Y.; Shu, Y.; Liu, P. miR-1271 Regulates Cisplatin Resistance of Human Gastric Cancer Cell Lines by Targeting IGF1R, IRS1, mTOR, and BCL2. Anticancer. Agents Med. Chem. 2014.
- Riquelme, I.; Letelier, P.; Riffo-Campos, A.L.; Brebi, P.; Roa, J.C. Emerging role of mirnas in the drug resistance of gastric cancer. Int. J. Mol. Sci. 2016, 17, 424.
- Denduluri, S.K.; Idowu, O.; Wang, Z.; Liao, Z.; Yan, Z.; Mohammed, M.K.; Ye, J.; Wei, Q.; Wang, J.; Zhao, L.; et al. Insulin-like growth factor (IGF) signaling intumorigenesis and the development ofcancer drug resistance. Genes Dis. 2015, 2, 13–25.
- Tao, Y.; Yang, S.; Wu, Y.; Fang, X.; Wang, Y.; Song, Y.; Han, T. MicroRNA-216a inhibits the metastasis of gastric cancer cells by targeting JAK2/STAT3-mediated EMT process. Oncotarget 2017, 8, 88870–88881.
- Song, H.; Shi, L.; Xu, Y.; Xu, T.; Fan, R.; Cao, M.; Xu, W.; Song, J. BRD4 promotes the stemness of gastric cancer cells via attenuating miR-216a-3p-mediated inhibition of Wnt/β-catenin signaling. Eur. J. Pharmacol. 2019.
- Wu, Y.; Zhang, J.; Zheng, Y.; Ma, C.; Liu, X.E.; Sun, X. miR-216a-3p inhibits the proliferation, migration, and invasion of human gastric cancer cells via targeting RUNX1 and activating the NF-κB signaling pathway. Oncol. Res. 2018.
- Wang, S.H.; Li, X.; Zhou, L.S.; Cao, Z.W.; Shi, C.; Zhou, C.Z.; Wen, Y.G.; Shen, Y.; Li, J.K. MicroRNA-148a suppresses human gastric cancer cell metastasis by reversing epithelial-to-mesenchymal transition. Tumor Biol. 2013.
- Zheng, B.; Liang, L.; Wang, C.; Huang, S.; Cao, X.; Zha, R.; Liu, L.; Jia, D.; Tian, Q.; Wu, J.; et al. MicroRNA-148a suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clin. Cancer Res. 2011.
- Li, B.; Wang, W.; Li, Z.; Chen, Z.; Zhi, X.; Xu, J.; Li, Q.; Wang, L.; Huang, X.; Wang, L.; et al. MicroRNA-148a-3p enhances cisplatin cytotoxicity in gastric cancer through mitochondrial fission induction and cyto-protective autophagy suppression. Cancer Lett. 2017.
- Li, Y.; Yan, X.; Shi, J.; He, Y.; Xu, J.; Lin, L.; Chen, W.; Lin, X.; Lin, X. Aberrantly expressed miR-188-5p promotes gastric cancer metastasis by activating Wnt/β-catenin signaling. BMC Cancer 2019.
- Yuan, J.; Tan, L.; Yin, Z.; Zhu, W.; Tao, K.; Wang, G.; Shi, W.; Gao, J. MIR17HG-miR-18a/19a axis, regulated by interferon regulatory factor-1, promotes gastric cancer metastasis via Wnt/β-catenin signalling. Cell Death Dis. 2019.
- Sun, G.L.; Li, Z.; Wang, W.Z.; Chen, Z.; Zhang, L.; Li, Q.; Wei, S.; Li, B.W.; Xu, J.H.; Chen, L.; et al. miR-324-3p promotes gastric cancer development by activating Smad4-mediated Wnt/beta-catenin signaling pathway. J. Gastroenterol. 2018.
- Zhi, X.; Tao, J.; Xiang, G.; Cao, H.; Liu, Z.; Yang, K.; Lv, C.; Ni, S. APRIL induces cisplatin resistance in gastric cancer cells via activation of the NF-κB pathway. Cell. Physiol. Biochem. 2015.
- Zhang, L.; Lei, J.; Fang, Z.L.; Xiong, J.P. MiR-128b is down-regulated in gastric cancer and negatively regulates tumour cell viability by targeting PDK1/Akt/NF-κB axis. J. Biosci. 2016.
- Tian, L.; Zhao, Z.; Xie, L.; Zhu, J.P. MiR-361-5p suppresses chemoresistance of gastric cancer cells by targeting FOXM1 via the PI3K/Akt/mTOR pathway. Oncotarget 2018.
- Medici, D.; Hay, E.D.; Olsen, B.R. Snail and slug promote epithelial-mesenchymal transition through β-catenin-T-cell factor-4-dependent expression of transforming growth factor-β3. Mol. Biol. Cell 2008.
- Xie, T.; Wu, D.; Li, S.; Li, X.; Wang, L.; Lu, Y.; Song, Q.; Sun, X.; Wang, X. Microrna-582 potentiates liver and lung metastasis of gastric carcinoma cells through the foxo3-mediated pi3k/akt/snail pathway. Cancer Manag. Res. 2020.
- Zhu, H.; Wu, Y.; Kang, M.; Zhang, B. MiR-877 suppresses gastric cancer progression by downregulating AQP3. J. Int. Med. Res. 2020.
- He, Y.; Ge, Y.; Jiang, M.; Zhou, J.; Luo, D.; Fan, H.; Shi, L.; Lin, L.; Yang, L. MiR-592 Promotes Gastric Cancer Proliferation, Migration, and Invasion Through the PI3K/AKT and MAPK/ERK Signaling Pathways by Targeting Spry2. Cell. Physiol. Biochem. 2018.
- Wang, J.; Wen, T.; Li, Z.; Che, X.; Gong, L.; Yang, X.; Zhang, J.; Tang, H.; He, L.; Qu, X.; et al. MicroRNA-1224 inhibits tumor metastasis in intestinal-type gastric cancer by directly targeting FAK. Front. Oncol. 2019.
- [39]
- Guan, H.; Li, W.; Li, Y.; Wang, J.; Li, Y.; Tang, Y.; Lu, S. MicroRNA-93 promotes proliferation and metastasis of gastric cancer via targeting TIMP2. PLoS ONE 2017.
- Cha, Y.; He, Y.; Ouyang, K.; Xiong, H.; Li, J.; Yuan, X. microRNA-140-5p suppresses cell proliferation and invasion in gastric cancer by targeting WNT1 in the WNT/β-catenin signaling pathway. Oncol. Lett. 2018, 16, 6369–6376.
- Zheng, L.; Qi, T.; Yang, D.; Qi, M.; Li, D.; Xiang, X.; Huang, K.; Tong, Q. microRNA-9 Suppresses the Proliferation, Invasion and Metastasis of Gastric Cancer Cells through Targeting Cyclin D1 and Ets1. PLoS ONE 2013.
- Wang, N.; Wang, L.; Yang, Y.; Gong, L.; Xiao, B.; Liu, X. A serum exosomal microRNA panel as a potential biomarker test for gastric cancer. Biochem. Biophys. Res. Commun. 2017.
- Virgilio, E.; Giarnieri, E.; Giovagnoli, M.R.; Montagnini, M.; Proietti, A.; D’Urso, R.; Mercantini, P.; Balducci, G.; Cavallini, M. Gastric juice MicroRNAs as potential biomarkers for screening gastric cancer: A systematic review. Anticancer Res. 2018, 38, 613–616.
- Yin, L.; Xu, G.; Zhu, Y.; Wang, Y. Expression of miR-23a and miR-135 and tumor markers in gastric cancer patients and the significance in diagnosis. Oncol. Lett. 2019, 18, 5853–5858.
- Dong, X.; Liu, Y. Expression and significance of miR-24 and miR-101 in patients with advanced gastric cancer. Oncol. Lett. 2018, 16, 5769–5774.
- Han, T.S.; Hur, K.; Xu, G.; Choi, B.; Okugawa, Y.; Toiyama, Y.; Oshima, H.; Oshima, M.; Lee, H.J.; Kim, V.N.; et al. MicroRNA-29c mediates initiation of gastric carcinogenesis by directly targeting ITGB1. Gut 2015.
- Cao, J.M.; Li, G.Z.; Han, M.; Xu, H.L.; Huang, K.M. MiR-30c-5p suppresses migration, invasion and epithelial to mesenchymal transition of gastric cancer via targeting MTA1. Biomed. Pharmacother. 2017.
- Kipkeeva, F.M.; Muzaffarova, Т.А.; Nikulin, M.P.; Apanovich, P.V.; Narimanov, M.N.; Malikhova, O.A.; Kushlinskii, N.E.; Stilidi, I.S.; Karpukhin, A.V. A Group of miRNA as Candidates for Prognostic Biomarkers of Gastric Cancer Metastasis. Bull. Exp. Biol. Med. 2020, 169, 77–80.
- Zeng, W.; Zhang, S.; Yang, L.; Wei, W.; Gao, J.; Guo, N.; Wu, F. Serum miR-101-3p combined with pepsinogen contributes to the early diagnosis of gastric cancer. BMC Med. Genet. 2020.
- Peng, Q.; Shen, Y.; Lin, K.; Zou, L.; Shen, Y.; Zhu, Y. Comprehensive and integrative analysis identifies microRNA-106 as a novel non-invasive biomarker for detection of gastric cancer. J. Transl. Med. 2018.
- Hwang, J.; Min, B.H.; Jang, J.; Kang, S.Y.; Bae, H.; Jang, S.S.; Kim, J.I.; Kim, K.M. MicroRNA Expression Profiles in Gastric Carcinogenesis. Sci. Rep. 2018, 8, 1–8.
- Link, A.; Schirrmeister, W.; Langner, C.; Varbanova, M.; Bornschein, J.; Wex, T.; Malfertheiner, P. Differential expression of microRNAs in preneoplastic gastric mucosa. Sci. Rep. 2015.
- Lu, Q.; Chen, Y.; Sun, D.; Wang, S.; Ding, K.; Liu, M.; Zhang, Y.; Miao, Y.; Liu, H.; Zhou, F. MicroRNA-181a functions as an oncogene in gastric cancer by targeting caprin-1. Front. Pharmacol. 2019.
- Li, Z.; Guo, Q.; Lu, Y.; Tian, T. Increased expression of miR-181d is associated with poor prognosis and tumor progression of gastric cancer. Cancer Biomark. 2019, 26, 353–360.
- Tsai, M.M.; Wang, C.S.; Tsai, C.Y.; Huang, C.G.; Lee, K.F.; Huang, H.W.; Lin, Y.H.; Chi, H.C.; Kuo, L.M.; Lu, P.H.; et al. Circulating microRNA-196a/b are novel biomarkers associated with metastatic gastric cancer. Eur. J. Cancer 2016.
- Liao, Z.; Li, Y.; Zhou, Y.; Huang, Q.; Dong, J. MicroRNA-197 inhibits gastric cancer progression by directly targeting metadherin. Mol. Med. Rep. 2018.
- Huang, Z.S.; Guo, X.W.; Zhang, G.; Liang, L.X.; Nong, B. The diagnostic and prognostic value of miR-200c in gastric cancer: A meta-analysis. Dis. Markers 2019.
- Ma, B.; Ma, J.; Yang, Y.; He, X.; Pan, X.; Wang, Z.; Qian, Y. Effects of mIR-330-3P on invasion, migration and EMT of gastric cancer cells by targeting PRRX1-mediated Wnt/β-catenin signaling pathway. OncoTargets Ther. 2020, 13, 3411–3423.
- Tian, L.; Zhao, Z.; Xie, L.; Zhu, J.P. MiR-361-5p inhibits the mobility of gastric cancer cells through suppressing epithelial-mesenchymal transition via the Wnt/β-catenin pathway. Gene 2018, 675, 102–109.
- Zhang, C.; Liang, Y.; Ma, M.H.; Wu, K.Z.; Zhang, C.D.; Dai, D.Q. Downregulation of microRNA-376a in gastric cancer and association with poor prognosis. Cell. Physiol. Biochem. 2018, 51, 2010–2018.
- Xu, M.; Qin, S.; Cao, F.; Ding, S.; Li, M. MicroRNA-379 inhibits metastasis and epithelial-mesenchymal transition via targeting FAK/AKT signaling in gastric cancer. Int. J. Oncol. 2017, 51, 867–876.
- Ge, X.; Liu, X.; Lin, F.; Li, P.; Liu, K.; Geng, R.; Dai, C.; Lin, Y.; Tang, W.; Wu, Z.; et al. MicroRNA-421 regulated by HIF-1α promotes metastasis, inhibits apoptosis, and induces cisplatin resistance by targeting E-cadherin and caspase-3 in gastric cancer. Oncotarget 2016.
- Chen, J.; Wu, L.; Sun, Y.; Yin, Q.; Chen, X.; Liang, S.; Meng, Q.; Long, H.; Li, F.; Luo, C.; et al. MIR-421 in plasma as a potential diagnostic biomarker for precancerous gastric lesions and early gastric cancer. PeerJ 2019.
- Li, Y.; Liu, Y.; Yao, J.; Li, R.; Fan, X. Downregulation of miR-484 is associated with poor prognosis and tumor progression of gastric cancer. Diagn. Pathol. 2020, 15, 1–7.
- Cai, H.; Lin, H.; Cao, W.; Sun, J.; Huang, Y.; Fang, Y. Downregulation of miR-519a Predicts Poor Prognosis and Contributes to Tumor Progression in Gastric Cancer. Oncol. Res. Treat. 2020, 43, 19–26.
- Su, H.; Ren, F.; Jiang, H.; Chen, Y.; Fan, X. Upregulation of microRNA-520a-3p inhibits the proliferation, migration and invasion via spindle and kinetochore associated 2 in gastric cancer. Oncol. Lett. 2019.
- Feng, X.; Zhu, M.; Liao, B.; Tian, T.; Li, M.; Wang, Z.; Chen, G. Upregulation of miR-552 Predicts Unfavorable Prognosis of Gastric Cancer and Promotes the Proliferation, Migration, and Invasion of Gastric Cancer Cells. Oncol. Res. Treat. 2020, 43, 103–110.
- Hu, L.; Wu, H.; Wan, X.; Liu, L.; He, Y.; Zhu, L.; Liu, S.; Yao, H.; Zhu, Z. MicroRNA-585 suppresses tumor proliferation and migration in gastric cancer by directly targeting MAPK1. Biochem. Biophys. Res. Commun. 2018.
- Min, C.; Zhang, A.; Qin, J. Increased expression of miR-601 is associated with poor prognosis and tumor progression of gastric cancer. Diagn. Pathol. 2019, 14, 1–7.
- Zheng, H.; Zhang, F.; Lin, X.; Huang, C.; Zhang, Y.; Li, Y.; Lin, J.; Chen, W.; Lin, X. MicroRNA-1225-5p inhibits proliferation and metastasis of gastric carcinoma through repressing insulin receptor substrate-1 and activation of β-catenin signaling. Oncotarget 2016.
- An, J.X.; Ma, Z.S.; Ma, M.H.; Shao, S.; Cao, F.L.; Dai, D.Q. MiR-1236-3p serves as a new diagnostic and prognostic biomarker for gastric cancer. Cancer Biomark. 2019, 25, 127–132.
- Shang, Y.; Feng, B.; Zhou, L.; Ren, G.; Zhang, Z.; Fan, X.; Sun, Y.; Luo, G.; Liang, J.; Wu, K.; et al. The miR27b-CCNG1-P53-miR-508-5p axis regulates multidrug resistance of gastric cancer. Oncotarget 2016.
- Zhang, J.X.; Xu, Y.; Gao, Y.; Chen, C.; Zheng, Z.S.; Yun, M.; Weng, H.W.; Xie, D.; Ye, S. Decreased expression of miR-939 contributes to chemoresistance and metastasis of gastric cancer via dysregulation of SLC34A2 and Raf/MEK/ERK pathway. Mol. Cancer 2017.
- Zhao, R.; Zhao, L.; Xu, X.U.; Xu, H. Analysis of microRNA expression profiles reveals a 5-microRNA prognostic signature for predicting overall survival time in patients with gastric adenocarcinoma. Oncol. Rep. 2019.
- Jiang, X.; Wang, W.; Yang, Y.; Du, L.; Yang, X.; Zheng, G.; Duan, W.; Wang, R.; Zhang, X.; Wang, L. Identification of circulating microRNA signatures as potential noninvasive biomarkers for prediction and prognosis of lymph node metastasis in gastric cancer. Oncotarget 2017, 8, 65132–65142.
- Wang, J.; Zhang, H.; Zhou, X.; Wang, T.; Zhang, J.; Zhu, W.; Zhu, H.; Cheng, W. Five serum-based miRNAs were identified as potential diagnostic biomarkers in gastric cardia adenocarcinoma. Cancer Biomark. 2018.
- Huang, Z.; Zhu, D.; Wu, L.; He, M.; Zhou, X.; Zhang, L.; Zhang, H.; Wang, W.; Zhu, J.; Cheng, W.; et al. Six serum-based miRNAs as potential diagnostic biomarkers for gastric cancer. Cancer Epidemiol. Biomark. Prev. 2017.
- Link, A.; Kupcinskas, J. MicroRNAs as non-invasive diagnostic biomarkers for gastric cancer: Current insights and future perspectives. World J. Gastroenterol. 2018, 24, 3313–3329.
- Rekker, K.; Saare, M.; Roost, A.M.; Kaart, T.; Sõritsa, D.; Karro, H.; Sõritsa, A.; Simón, C.; Salumets, A.; Peters, M. Circulating miR-200-family micro-RNAs have altered plasma levels in patients with endometriosis and vary with blood collection time. Fertil. Steril. 2015.
- Suryawanshi, S.; Vlad, A.M.; Lin, H.M.; Mantia-Smaldone, G.; Laskey, R.; Lee, M.; Lin, Y.; Donnellan, N.; Klein-Patel, M.; Lee, T.; et al. Plasma MicroRNAs as novel biomarkers for endometriosis and endometriosis-associated ovarian cancer. Clin. Cancer Res. 2013.
- Jia, S.Z.; Yang, Y.; Lang, J.; Sun, P.; Leng, J. Plasma miR-17-5p, miR-20a and miR-22 are down-regulated in women with endometriosis. Hum. Reprod. 2013, 28, 322–330.
- Gao, J.; Yang, S.; Wang, K.; Zhong, Q.; Ma, A.; Pan, X. Plasma miR-126 and miR-143 as Potential Novel Biomarkers for Cerebral Atherosclerosis. J. Stroke Cerebrovasc. Dis. 2019.
- Zhou, J.; Chen, Q. Poor expression of microRNA-135b results in the inhibition of cisplatin resistance and proliferation and induces the apoptosis of gastric cancer cells through MST1-mediated MAPK signaling pathway. FASEB J. 2019.
- Wang, Q.; Cao, T.; Guo, K.; Zhou, Y.; Liu, H.; Pan, Y.; Hou, Q.; Nie, Y.; Fan, D.; Lu, Y.; et al. Regulation of Integrin Subunit Alpha 2 by miR-135b-5p Modulates Chemoresistance in Gastric Cancer. Front. Oncol. 2020.
- Liu, R.; Zhang, C.; Hu, Z.; Li, G.; Wang, C.; Yang, C.; Huang, D.; Chen, X.; Zhang, H.; Zhuang, R.; et al. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur. J. Cancer 2011.
- Zhu, C.; Ren, C.; Han, J.; Ding, Y.; Du, J.; Dai, N.; Dai, J.; Ma, H.; Hu, Z.; Shen, H.; et al. A five-microRNA panel in plasma was identified as potential biomarker for early detection of gastric cancer. Br. J. Cancer 2014, 2291–2299.
- Kim, D.N.; Chae, H.-S.; Oh, S.T.; Kang, J.-H.; Park, C.H.; Park, W.S.; Takada, K.; Lee, J.M.; Lee, W.-K.; Lee, S.K. Expression of Viral MicroRNAs in Epstein-Barr Virus-Associated Gastric Carcinoma. J. Virol. 2007.
- Treece, A.L.; Duncan, D.L.; Tang, W.; Elmore, S.; Morgan, D.R.; Dominguez, R.L.; Speck, O.; Meyers, M.O.; Gulley, M.L. Gastric adenocarcinoma microRNA profiles in fixed tissue and in plasma reveal cancer-associated and Epstein-Barr virus-related expression patterns. Lab. Investig. 2016.
