Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Exosomes are nanometer-sized vesicles released by different cells that are important in the normal functioning of the body. In cancer, exosomes have been found to promote tumor growth and metastasis by carrying functional biomolecules and acting on different target sites in the body. Understanding the mechanism by which cancers modulate exosome secretion is crucial to studying cancer biology and developing new therapeutic approaches.
- regulation
- cancer
- endocytosis
1. Mechanisms of Exosome Biogenesis
The pathway of exosome biogenesis is the most well-studied among the subsets of extracellular vesicles (EVs). Given their functional importance and the translational applications focused on exosomes exclusively, a mechanistic understanding of exosome biogenesis and absorption is necessary to form functional hypotheses.
In general, exosome biogenesis is preceded by endocytosis, resulting in the formation of early endosomes. Several mechanisms cause the early endosome to incorporate cargo into intraluminal vesicles (ILVs), forming multivesicular bodies (MVBs) or late endosomes [1]. During maturation, MVBs may import cargoes from other organelles, including the trans-Golgi network (TGN), endoplasmic reticulum (ER) and other cytosolic compartments [2][3], further modifying the composition of ILVs. MVBs may fuse with lysosomes, resulting in degradation [4]. Alternatively, MVBs are transported to the plasma membrane, followed by membrane fusion, releasing ILVs as exosomes. The exosomes travel across extracellular space and reach a recipient cell, where several factors mediate the interaction between the exosome and the recipient cell [5]. Internalized exosomes are incorporated into early endosomes following the typical endosomal pathway and may be released again through recycling endosomes [6]. The overview of exosome biogenesis is depicted in Figure 1.
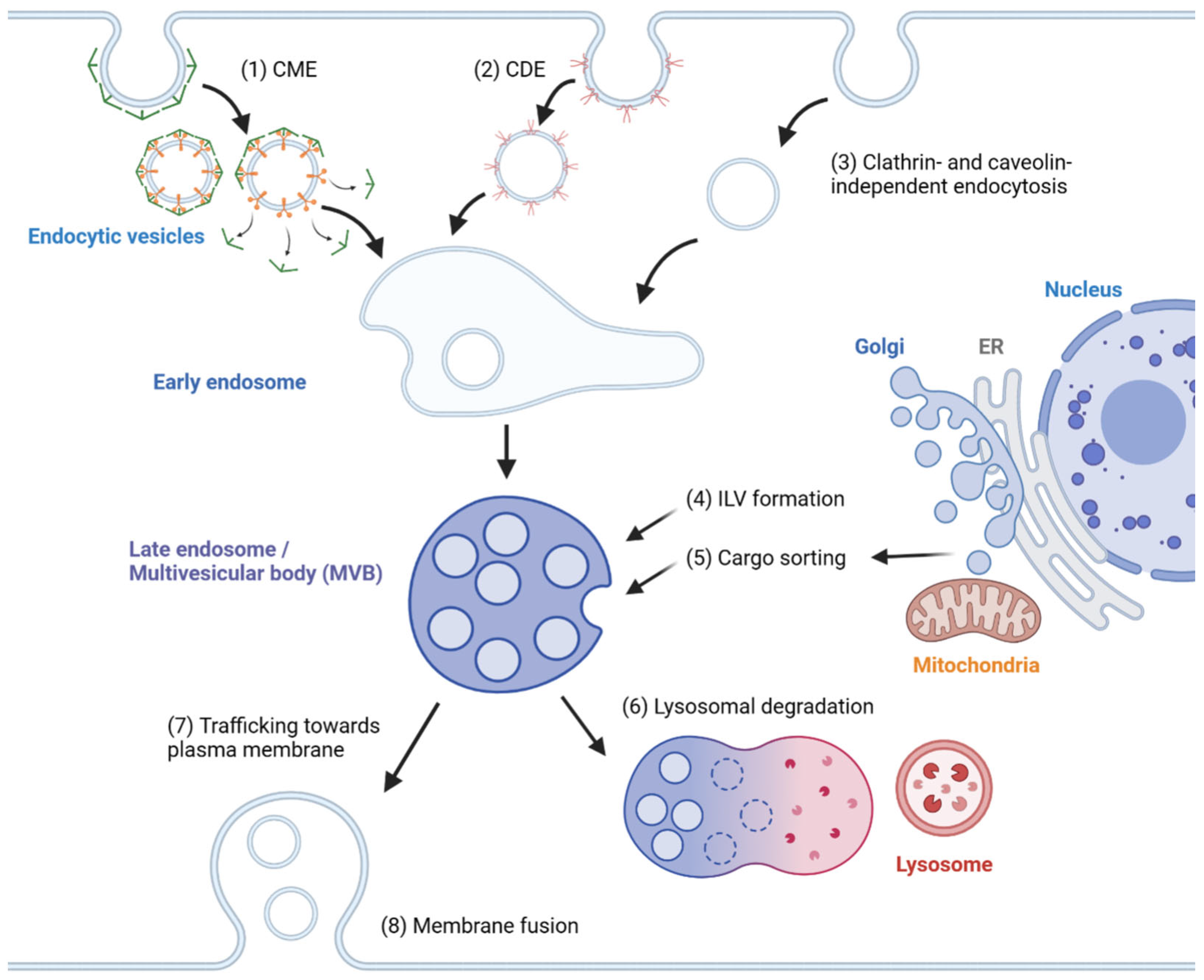
Figure 1. Overview of exosome biogenesis. Endocytic vesicles are generated by (1) clathrin-mediated endocytosis (CME), (2) caveolin-dependent endocytosis (CDE) and (3) clathrin- and caveolin-independent endocytosis. After the fusion of endocytic vesicles to form early endosomes, (4) ILVs are generated by ESCRT-dependent and ESCRT-independent pathways, resulting in the formation of multivesicular bodies (MVBs). (5) Cargoes are sorted into ILVs from various organelles, including the trans-Golgi network (TGN), endoplasmic reticulum (ER) and mitochondria. (6) MVBs can then fuse with lysosomes and undergo lysosomal degradation. (7) Alternatively, MVBs are trafficked towards the plasma membrane and (8) undergo membrane fusion for the extracellular release of ILVs as exosomes.
To summarize, exosome biogenesis consists of endosome formation, MVB formation, cargo sorting and extracellular release, while exosome absorption consists of targeting to the recipient cell, internalization and re-release. Each step is regulated by molecular pathways and environmental factors, leading to changes in the amount, composition, and eventually the dynamics of exosomes in a biological system. Despite sharing these common steps, exosomes isolated from biofluids are still highly heterogeneous due to cell-type-specific regulation of biogenesis and absorption, as well as the intracellular heterogeneity of MVBs and ILVs, the mechanisms for which will be highlighted in this section.
2. Endosome Formation
Early endosomes are the precursors of MVBs. Although a crucial condition for subsequent steps, it has yet to be sufficiently characterized in the context of exosome biogenesis. There are multiple pathways of endocytosis, including clathrin-mediated endocytosis (CME), caveolin-dependent endocytosis (CDE) and clathrin- and caveolin-independent endocytosis [7][8]. The mechanism for CME has been reviewed in detail elsewhere [7]. In short, a FCH domain only (FCHO) initiation complex is formed on the plasma membrane, forming a clathrin-coated pit. These proteins, along with adaptor protein 2 (AP2) and cargo-specific adaptor proteins, mediate cargo selection, followed by clathrin coat assembly and dynamin-mediated vesicle scission. The clathrin coat is finally disassembled by the ATPase heat shock cognate 70 (HSC70) and auxilin [7]. Other pathways include CDE, the clathrin-independent carriers/GPI-AP-enriched early endosomal compartments (CLIC/GEEC) pathway and the ARF6-associated pathway. Though the detailed mechanism of these pathways are not well defined, the reorganization of the actin cytoskeleton appears to play a central role in endosome formation, especially in pathways independent of clathrin or caveolin coating [8].
3. MVB Formation and Cargo Encapsulation
The endosomal sorting complex required for transport (ESCRT)-dependent pathway has been well studied for its involvement in ILV generation. The ESCRT machinery comprises ESCRT-0, -I, -II and -III complexes and Vps4 [9]. ESCRT-0 binds to ubiquitinated cargoes via the hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs) subunit, while binding to phosphatidylinositol-3-phosphate (PI3P) on endosomal membranes, resulting in cargo enrichment on the limiting membrane of endosomes [9][10]. ESCRT-I is recruited to ESCRT-0 via the interaction of the ESCRT-I subunit, tumor susceptibility gene 101 (Tsg101) protein, with Hrs. This is followed by ESCRT-II binding to ESCRT-I. Together, they promote the inward budding of the endosomal membrane. Recruited by ESCRT-II, the ESCRT-III subunits Vps20, Snf7, Vps2 and Vps24 are sequentially assembled and promote the growth of the Snf7 polymer around the neck of the inward bud, driving further deformation, membrane fission and the formation of ILV [11]. The ESCRT-III subunit Did2 then replaces Vps24 and activates ATP hydrolysis by Vps4, resulting in the cleavage and disassembly of the ESCRT-III complex [11].
Numerous ESCRT-associated machinery cooperate with ESCRT in the formation of ILVs. Non-canonical ESCRT-dependent pathways, such as the Syndecan-Syntenin-Alix pathway and the his domain protein tyrosine phosphatase (HD-PTP) pathway also recruit ESCRT-III and VPS4 for the budding and scission of ILVs. The Syndecan–Syntenin–Alix pathway is initiated by the presence of the ubiquitous transmembrane protein Syndecan, recruiting Syntenin, which in turn binds Alix [12]. Proteins that are recognized by Syndecan, Syntenin or Alix are preferentially sorted into exosomes, such as fibroblast growth factor receptor (FGFR), lysyl-tRNA synthetase and tetraspanins, including CD9, CD63 and CD81 [12][13][14]. The phosphorylation of Syntenin by proteins and the cleavage by heparinase regulates the activity of the Alix-dependent pathway [15][16][17]. The HD-PTP pathway is initiated by the binding of HD-PTP to ESCRT-0, recruiting ESCRT-III and VPS4, thus re-entering the typical ESCRT-dependent pathway [18]. Components of the ESCRT pathway may interact with cargoes to promote their exosomal sorting; namely, the interaction of BAG6 and beta-catenin with Tsg101 and ESCRT-III, respectively, mediates their exosomal secretion [19][20].
The ESCRT-independent pathway involves a combination of lipid raft components, which together regulate the budding and formation of ILVs [21][22]. The neutral sphingomyelinase 2 (nSMase2)-ceramide pathway involves the conversion of sphingomyelin to ceramide by nSMase2, which allows ceramide to self-associate into raft-like macrodomains, leading to a negative membrane curvature that favors inward budding [23]. Microtubule-associated protein 1 light chain 3 (LC3), a well-known marker of autophagy, is located on the limiting membrane of MVBs and recruits the FAN protein, which promotes nSMase2-mediated ceramide production [24][25].
Other lipid raft components, such as cholesterol, caveolin-1 and flotillins, have been linked to exosome secretion and the ILV sorting of cargoes [26][27][28][29]. Proteins that promote cholesterol transfer to MVBs, such as ORP1L and STARD3, positively regulate the formation of ILVs [27][30]. While the precise mechanism of how cholesterol mediates ILV formation is unknown, cholesterol is found to promote flotillin 2 secretion in oligodendroglial cells, and flotillin-1 knock-down inhibits caveolin-1 secretion in PC-3 cells [29][31]. Thus, it is attractive to hypothesize whether cholesterol, caveolin-1 and flotillins constitute an alternative pathway of ILV formation, and whether they are related to ESCRT or ESCRT-independent machinery. However, cholesterol can have different effects on exosome release depending on the cell type; therefore, mechanistic studies using consistent cell types are preferred before a pathway can be confidently established.
Tetraspanins are integral membrane proteins that contribute to protein scaffolding in tetraspanin-enriched microdomains, which mediate adhesion and signal transduction [32]. CD63 has been found to interact with Apolipoprotein E to promote ILV formation in melanocytes independently of ESCRT and ceramide [33]. CD63 has also been used to promote reporter cargo loading in engineered exosomes [34]. Tetraspanin-6 (TSPN6) promotes exosomal secretion in HEK293 cells by interacting with Syntenin, likely co-opting the Syntenin–Syndecan–Alix pathway [17][35], though conflicting evidence suggests that TSPN6 inhibits exosome release in MCF-7 cells due to directing cargoes towards lysosomal degradation [36]. As common exosome biomarkers, CD9 and CD81 are enriched on exosomes, but their mechanisms are not well-defined [37]. While CD9 interacts with and mediates the exosomal secretion of CD10 [38], specific CD81 ligands appear to be abundant on tetraspanin-enriched microdomains [39], suggesting CD81 involvement in cargo sorting. Generally, tetraspanins mediate cargo loading into ILVs via molecular interaction, but their role in the formation of ILV, or its relevance or necessity for the ESCRT and ESCRT-independent pathways, has yet to be clearly investigated.
4. Cargo Sorting to MVBs
Cargo sorting modulates the composition of ILVs and eventually exosomes. Exosomal cargo include proteins, amino acids, lipids, metabolites, DNA and RNAs, including messenger RNA (mRNA), microRNA (miRNA), long non-coding RNA (lncRNA), circular RNA (circRNA) and PIWI-interacting RNA (piRNA) [4][40]. The state of the donor cell can impact the proteomic and lipidomic profiles of the exosomal cargo [41]. As mentioned, protein cargoes are sorted into ILVs by interacting with components of the ESCRT-dependent or ESCRT-independent pathways, such as ESCRT-III, Alix, Syntenin-1 and ceramides [12][13][14][19][20][41]. In addition, Hsp90 alpha has been found to sort into ILVs by interacting with Rab coupling protein (RCP) [42] (Figure 2).
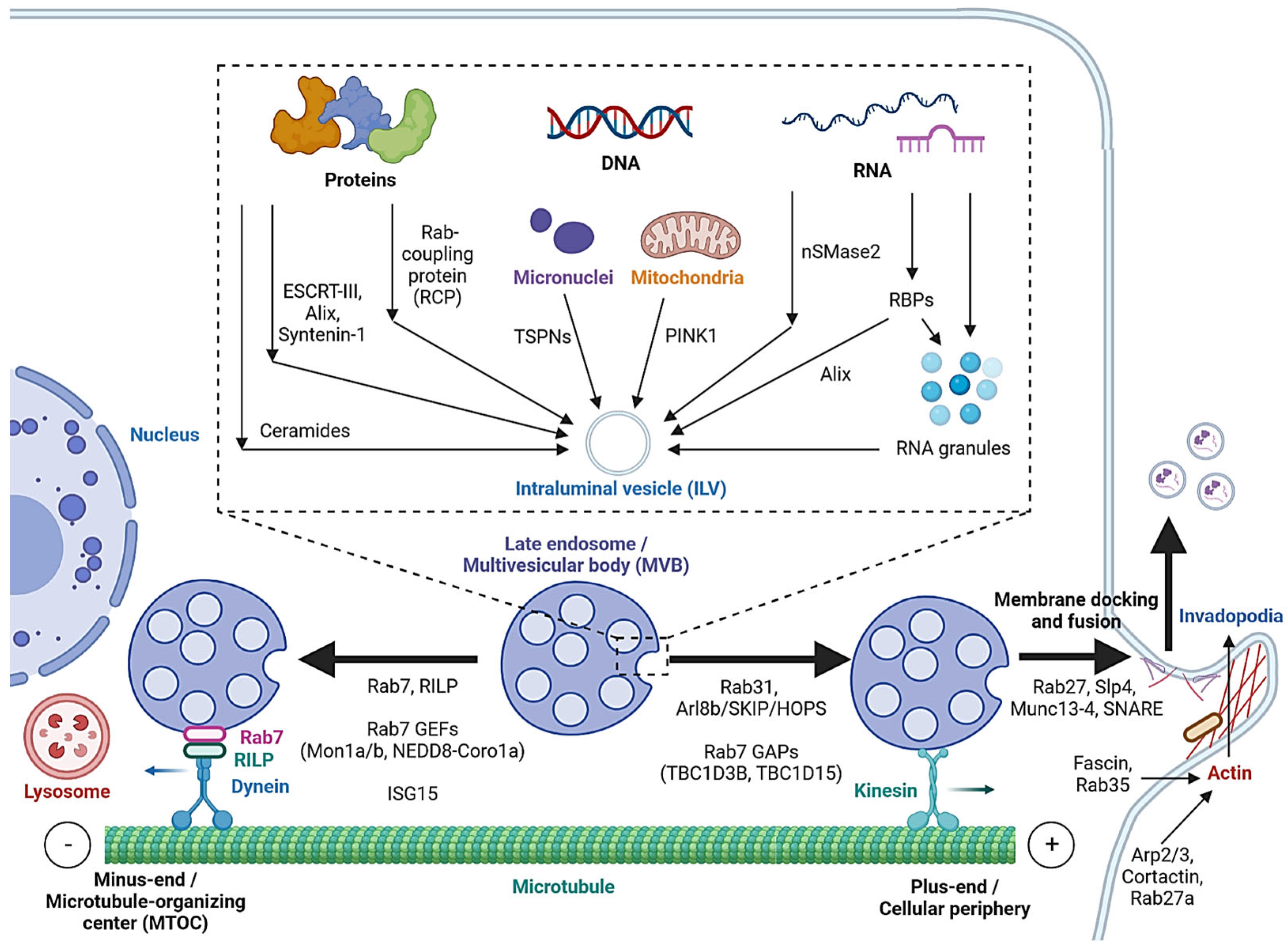
Figure 2. Cargo sorting mechanisms and the regulation of MVB trafficking and fusion with the cell membrane. Proteins are sorted into ILVs by associating with ESCRT-dependent and ESCRT-independent machinery and Rab coupling protein (RCP). Tetraspanins and PINK1 are involved in the loading of genomic DNA (gDNA) and mitochondrial DNA (mtDNA), respectively. RNA loading is regulated by nSMase2 and RNA-binding proteins (RBPs). Alix binds to the RBP Ago2 and promotes miRNA loading. RNA granules formed by liquid–liquid phase separation (LLPS) contain RNAs and RBPs and promote miRNA, lncRNA and circRNA loading. MVBs can be fated towards lysosomal degradation by translocation towards the microtubule minus-end or the perinuclear region. Alternatively, they are fated towards secretion by translocation towards the plus-end or the cellular periphery. Actin polymerization at invadopodia stabilizes MVB docking sites. MVB fusion is mediated by SNARE proteins, which are regulated by Rab27 and its effectors, Slp4 and Munc13-4.
While cell-free DNA (cfDNA) originates from various sources, a significant portion is localized on the surface or in the lumen of EVs [43]. Tetraspanins directly interact with cancer cell micronuclei, resulting in genomic DNA (gDNA) loading to exosomes [44]. Exosomal release also serves as a mechanism for removing harmful gDNA from the cytoplasm of healthy cells [45], whereas mitochondrial DNA (mtDNA) loading is initiated by mitochondrial damage, activating PINK1, which promotes the interaction of mitochondria and MVBs in an autophagy-independent manner [46].
Various mechanisms have been studied for RNA loading. nSMase2, which is involved in the ESCRT-independent pathway, has been reported to regulate the exosomal loading of miRNA in cancer cells [47][48]. Substance P (SP)/Neurokinin-1 Receptor (NK1R) signaling also increases miRNA expression and exosomal loading in colonocytes [49]. In addition, a number of RNA-binding proteins (RBPs), such as heterogeneous nuclear ribonucleoproteins (hnRNPs), Argonaute 2 (AGO2), Y-Box-Binding Protein-1 (YBX-1), Serine and Arginine-Rich Splicing Factor (SRSF1) and Major Vault Protein (MVP), bind to specific miRNAs motifs to facilitate their sorting to exosomes [50][51][52]. Meanwhile, hnRNPs also mediate the sorting of lncRNAs [53][54], and ESCRT-II and hnRNPA2B1 have been found to mediate the sorting of circRNAs [55][56]. Alix, well-known for its role in the ESCRT-dependent pathway, was also demonstrated to enrich miRNA in EVs via binding to Ago2 and miRNA [57].
In cells, biomolecular condensates, such as RNA granules, are formed by liquid–liquid phase separation (LLPS) [58]. Mechanistically, RNAs and RBPs contribute to RNA granule formation, which in turn contains components such as hnRNPA2 that facilitate miRNA, lncRNA and circRNA sorting into exosomes [58][59]. YBX-1 also forms LLPS-mediated condensates to promote miRNA loading into exosomes [60]. In addition, post-transcriptional modifications of miRNA sequences, in particular 3′ end adenylation and 3′ end uridylation, favor the cellular retention and exosomal release of miR-2909, respectively [61].
5. Extracellular Release
MVB translocation and fusion with the plasma membrane are required for the release of ILVs as exosomes. It primarily involves SNARE proteins that are well-known for their role in mediating membrane fusion events [62]. Various upstream mechanisms regulate the structure of SNAREs in different cell types that favor the fusion of MVBs with the plasma membrane, namely, the Fas/Fap-1/caveolin-1 cascade regulates the formation of SNARE containing SNAP25 and VAMP5 in mesenchymal stem cells [63], while the lncRNA HOTAIR regulates the formation of SNARE containing SNAP23 and VAMP3 in hepatocellular carcinoma [64].
As mentioned, MVBs can be fated towards the degradative pathway or the secretory pathway, based on their eventual fusion with lysosomes or with the cell membrane. Cytoskeleton filaments are crucial for MVB transport and docking. Actin reorganization, invadopodia formation and cortactin promote exosome secretion [65][66]. Cortactin binds to Arp2/3 and to actin filaments, leading to the stabilization of MVB docking sites and promote exosome secretion [66]. Coordinately, Rab27a promotes MVB docking by preventing Coronin1b localization to invadopodia, antagonization of cortactin and actin disassembly [66]. Rab35 binds and localizes the actin-bundling protein Fascin to actin, which facilitates actin cross-linking [67]. Accordingly, impeding Cortactin and Fascin-1 reduced EV secretion by regulating invadopodia formation [68]. Microtubules, on the other hand, play a role in the MVB fate by enabling movement towards their minus-ends or the plus-ends [69]. Trafficking towards the plus-end or the cellular periphery promotes membrane docking and the secretory fate of MVBs, whereas the minus-end is anchored in the perinuclear microtubule-organizing center (MTOC) and promotes the lysosomal degradative fate of MVBs.
Rab7 regulates MVB movement along cytoskeleton microtubules. Cargo movement towards the plus-end and minus-end of microtubules is mediated by kinesin and the dynein–dynactin complex, respectively [70]. Rab7 binds to its trafficking adaptor protein, Rab-interacting lysosomal protein (RILP), to recruit the dynein motor complex, thus promoting the transport of Rab7-positive vesicles towards the perinuclear region [71]. Cleavage of RILP due to inflammasome activation inhibits Rab7 binding to dynein motor complex, leading to kinesin-mediated movement of Rab7-positive vesicles to the cell periphery [72]. Thus, Rab7 appears to direct MVBs towards degradation, and its regulation alters the fate of MVBs. Rab31 recruits the GTPase-activating protein (GAP) TBC1D2B, which inhibits Rab7, resulting in increased MVB membrane fusion and exosome secretion [73]. Arl8b, also present on endosomal membranes, initiates the Arl8b/SKIP/HOPS cascade to recruit another GAP TBC1D15, which inhibits Rab7 [69]. As opposed to GAPs, the guanine nucleotide exchange factors (GEFs) Mon1a/b and NEDD8-Coro1a activate Rab7 by converting it to its GTP-bound state, resulting in lysosomal targeting and reduced EV secretion [74][75]. Additionally, induction of ISGylation via interferon-stimulating gene 15 (ISG15) overexpression was found to induce MVB protein degradation and colocalization with lysosomes [76].
Rab GTPases also control the MVB distribution, membrane docking and SNARE-mediated membrane fusion. Slp4 and Munc13-4 are downstream effectors of Rab27 that bind to and promote the formation of SNARE proteins to mediate the fusion of MVBs with the plasma membrane [77][78][79]. GEFs that regulate Rab27a/b, such as the DENN domain-containing protein Rab-3GEP (MADD) and FAM45A, promote endosomal maturation [80][81], and the stabilization of Rab27a by KIBRA also contributes to exosome secretion [82]. Rab27b regulates the trafficking of MVBs towards the plasma membrane by its effector Slac2b [77]. Rab11 and Rab35 enhance exosomal secretion in leukemia and oligodendrocytes, respectively [83][84]. Calcium ions (Ca2+) is required for the Munc13-4- and Rab11-mediated pathways of exosome secretion [85][86].
In summary, cytoskeleton components, such as actin filaments and microtubules, are crucial in MVB trafficking, docking and release. These processes are regulated by Rabs, which are in turn controlled by their GEFs and GAPs. Further study of Rabs and their effectors may help uncover viable mechanisms that modulate the volume and composition of secreted exosomes.
This entry is adapted from the peer-reviewed paper 10.3390/cancers15071992
References
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228.
- Kwon, S.H.; Oh, S.; Nacke, M.; Mostov, K.E.; Lipschutz, J.H. Adaptor Protein CD2AP and L-type Lectin LMAN2 Regulate Exosome Cargo Protein Trafficking through the Golgi Complex. J. Biol. Chem. 2016, 291, 25462–25475.
- Di Mattia, T.; Tomasetto, C.; Alpy, F. Faraway, so close! Functions of Endoplasmic reticulum–Endosome contacts. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158490.
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, 6977.
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021, 19, 47.
- O’Brien, K.; Ughetto, S.; Mahjoum, S.; Nair, A.V.; Breakefield, X.O. Uptake, functionality, and re-release of extracellular vesicle-encapsulated cargo. Cell Rep. 2022, 39, 110651.
- McMahon, H.T.; Boucrot, E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2011, 12, 517–533.
- Mayor, S.; Parton, R.G.; Donaldson, J.G. Clathrin-independent pathways of endocytosis. Cold Spring Harb. Perspect. Biol. 2014, 6, 16758.
- Schmidt, O.; Teis, D. The ESCRT machinery. Curr. Biol. 2012, 22, R116–R120.
- Vietri, M.; Radulovic, M.; Stenmark, H. The many functions of ESCRTs. Nat. Rev. Mol. Cell Biol. 2020, 21, 25–42.
- Pfitzner, A.K.; Mercier, V.; Jiang, X.; Moser von Filseck, J.; Baum, B.; Šarić, A.; Roux, A. An ESCRT-III Polymerization Sequence Drives Membrane Deformation and Fission. Cell 2020, 182, 1140–1155.e1118.
- Baietti, M.F.; Zhang, Z.; Mortier, E.; Melchior, A.; Degeest, G.; Geeraerts, A.; Ivarsson, Y.; Depoortere, F.; Coomans, C.; Vermeiren, E.; et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012, 14, 677–685.
- Kim, S.B.; Kim, H.R.; Park, M.C.; Cho, S.; Goughnour, P.C.; Han, D.; Yoon, I.; Kim, Y.; Kang, T.; Song, E.; et al. Caspase-8 controls the secretion of inflammatory lysyl-tRNA synthetase in exosomes from cancer cells. J. Cell Biol. 2017, 216, 2201–2216.
- Larios, J.; Mercier, V.; Roux, A.; Gruenberg, J. ALIX- and ESCRT-III-dependent sorting of tetraspanins to exosomes. J. Cell Biol. 2020, 219, 4113.
- Zhang, Y.; Li, Y.; Liu, P.; Gong, D.; Zhou, H.; Li, W.; Zhang, H.; Zheng, W.; Xu, J.; Cheng, H.; et al. Phosphatase Shp2 regulates biogenesis of small extracellular vesicles by dephosphorylating Syntenin. J. Extracell. Vesicles 2021, 10, e12078.
- Hyenne, V.; Labouesse, M.; Goetz, J.G. The Small GTPase Ral orchestrates MVB biogenesis and exosome secretion. Small GTPases 2018, 9, 445–451.
- Roucourt, B.; Meeussen, S.; Bao, J.; Zimmermann, P.; David, G. Heparanase activates the syndecan-syntenin-ALIX exosome pathway. Cell Res. 2015, 25, 412–428.
- Kazan, J.M.; Desrochers, G.; Martin, C.E.; Jeong, H.; Kharitidi, D.; Apaja, P.M.; Roldan, A.; St Denis, N.; Gingras, A.C.; Lukacs, G.L.; et al. Endofin is required for HD-PTP and ESCRT-0 interdependent endosomal sorting of ubiquitinated transmembrane cargoes. iScience 2021, 24, 103274.
- Schuldner, M.; Dörsam, B.; Shatnyeva, O.; Reiners, K.S.; Kubarenko, A.; Hansen, H.P.; Finkernagel, F.; Roth, K.; Theurich, S.; Nist, A.; et al. Exosome-dependent immune surveillance at the metastatic niche requires BAG6 and CBP/p300-dependent acetylation of p53. Theranostics 2019, 9, 6047–6062.
- Han, Q.; Lv, L.; Wei, J.; Lei, X.; Lin, H.; Li, G.; Cao, J.; Xie, J.; Yang, W.; Wu, S.; et al. Vps4A mediates the localization and exosome release of β-catenin to inhibit epithelial-mesenchymal transition in hepatocellular carcinoma. Cancer Lett. 2019, 457, 47–59.
- Skotland, T.; Hessvik, N.P.; Sandvig, K.; Llorente, A. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J. Lipid. Res. 2019, 60, 9–18.
- Dawson, G. Isolation of Lipid Rafts (Detergent-Resistant Microdomains) and Comparison to Extracellular Vesicles (Exosomes). Methods Mol. Biol. 2021, 2187, 99–112.
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247.
- Leidal, A.M.; Huang, H.H.; Marsh, T.; Solvik, T.; Zhang, D.; Ye, J.; Kai, F.; Goldsmith, J.; Liu, J.Y.; Huang, Y.H.; et al. The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat. Cell Biol. 2020, 22, 187–199.
- Adam-Klages, S.; Adam, D.; Wiegmann, K.; Struve, S.; Kolanus, W.; Schneider-Mergener, J.; Krönke, M. FAN, a novel WD-repeat protein, couples the p55 TNF-receptor to neutral sphingomyelinase. Cell 1996, 86, 937–947.
- Han, Q.F.; Li, W.J.; Hu, K.S.; Gao, J.; Zhai, W.L.; Yang, J.H.; Zhang, S.J. Exosome biogenesis: Machinery, regulation, and therapeutic implications in cancer. Mol. Cancer 2022, 21, 207.
- Eden, E.R.; Sanchez-Heras, E.; Tsapara, A.; Sobota, A.; Levine, T.P.; Futter, C.E. Annexin A1 Tethers Membrane Contact Sites that Mediate ER to Endosome Cholesterol Transport. Dev. Cell 2016, 37, 473–483.
- Albacete-Albacete, L.; Navarro-Lérida, I.; López, J.A.; Martín-Padura, I.; Astudillo, A.M.; Ferrarini, A.; Van-Der-Heyden, M.; Balsinde, J.; Orend, G.; Vázquez, J.; et al. ECM deposition is driven by caveolin-1-dependent regulation of exosomal biogenesis and cargo sorting. J. Cell Biol. 2020, 219, 6178.
- Phuyal, S.; Hessvik, N.P.; Skotland, T.; Sandvig, K.; Llorente, A. Regulation of exosome release by glycosphingolipids and flotillins. FEBS J. 2014, 281, 2214–2227.
- Wilhelm, L.P.; Wendling, C.; Védie, B.; Kobayashi, T.; Chenard, M.P.; Tomasetto, C.; Drin, G.; Alpy, F. STARD3 mediates endoplasmic reticulum-to-endosome cholesterol transport at membrane contact sites. EMBO J. 2017, 36, 1412–1433.
- Strauss, K.; Goebel, C.; Runz, H.; Möbius, W.; Weiss, S.; Feussner, I.; Simons, M.; Schneider, A. Exosome secretion ameliorates lysosomal storage of cholesterol in Niemann-Pick type C disease. J. Biol. Chem. 2010, 285, 26279–26288.
- Kummer, D.; Steinbacher, T.; Schwietzer, M.F.; Thölmann, S.; Ebnet, K. Tetraspanins: Integrating cell surface receptors to functional microdomains in homeostasis and disease. Med. Microbiol. Immunol. 2020, 209, 397–405.
- van Niel, G.; Charrin, S.; Simoes, S.; Romao, M.; Rochin, L.; Saftig, P.; Marks, M.S.; Rubinstein, E.; Raposo, G. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev. Cell 2011, 21, 708–721.
- Bebelman, M.P.; Bun, P.; Huveneers, S.; van Niel, G.; Pegtel, D.M.; Verweij, F.J. Real-time imaging of multivesicular body-plasma membrane fusion to quantify exosome release from single cells. Nat. Protoc. 2020, 15, 102–121.
- Guix, F.X.; Sannerud, R.; Berditchevski, F.; Arranz, A.M.; Horré, K.; Snellinx, A.; Thathiah, A.; Saido, T.; Saito, T.; Rajesh, S.; et al. Tetraspanin 6: A pivotal protein of the multiple vesicular body determining exosome release and lysosomal degradation of amyloid precursor protein fragments. Mol. Neurodegener. 2017, 12, 25.
- Ghossoub, R.; Chéry, M.; Audebert, S.; Leblanc, R.; Egea-Jimenez, A.L.; Lembo, F.; Mammar, S.; Le Dez, F.; Camoin, L.; Borg, J.P.; et al. Tetraspanin-6 negatively regulates exosome production. Proc. Natl. Acad. Sci. USA 2020, 117, 5913–5922.
- Escola, J.M.; Kleijmeer, M.J.; Stoorvogel, W.; Griffith, J.M.; Yoshie, O.; Geuze, H.J. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 1998, 273, 20121–20127.
- Mazurov, D.; Barbashova, L.; Filatov, A. Tetraspanin protein CD9 interacts with metalloprotease CD10 and enhances its release via exosomes. FEBS J. 2013, 280, 1200–1213.
- Perez-Hernandez, D.; Gutiérrez-Vázquez, C.; Jorge, I.; López-Martín, S.; Ursa, A.; Sánchez-Madrid, F.; Vázquez, J.; Yáñez-Mó, M. The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J. Biol. Chem. 2013, 288, 11649–11661.
- Bobrie, A.; Colombo, M.; Raposo, G.; Théry, C. Exosome secretion: Molecular mechanisms and roles in immune responses. Traffic 2011, 12, 1659–1668.
- Ferreira, J.V.; da Rosa Soares, A.; Ramalho, J.; Máximo Carvalho, C.; Cardoso, M.H.; Pintado, P.; Carvalho, A.S.; Beck, H.C.; Matthiesen, R.; Zuzarte, M.; et al. LAMP2A regulates the loading of proteins into exosomes. Sci. Adv. 2022, 8, eabm1140.
- Zhang, S.; Wang, C.; Ma, B.; Xu, M.; Xu, S.; Liu, J.; Tian, Y.; Fu, Y.; Luo, Y. Mutant p53 Drives Cancer Metastasis via RCP-Mediated Hsp90α Secretion. Cell Rep. 2020, 32, 107879.
- Grabuschnig, S.; Bronkhorst, A.J.; Holdenrieder, S.; Rosales Rodriguez, I.; Schliep, K.P.; Schwendenwein, D.; Ungerer, V.; Sensen, C.W. Putative Origins of Cell-Free DNA in Humans: A Review of Active and Passive Nucleic Acid Release Mechanisms. Int. J. Mol. Sci. 2020, 21, 8062.
- Yokoi, A.; Villar-Prados, A.; Oliphint, P.A.; Zhang, J.; Song, X.; De Hoff, P.; Morey, R.; Liu, J.; Roszik, J.; Clise-Dwyer, K.; et al. Mechanisms of nuclear content loading to exosomes. Sci. Adv. 2019, 5, eaax8849.
- Takahashi, A.; Okada, R.; Nagao, K.; Kawamata, Y.; Hanyu, A.; Yoshimoto, S.; Takasugi, M.; Watanabe, S.; Kanemaki, M.T.; Obuse, C.; et al. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat. Commun. 2017, 8, 15287.
- Rabas, N.; Palmer, S.; Mitchell, L.; Ismail, S.; Gohlke, A.; Riley, J.S.; Tait, S.W.G.; Gammage, P.; Soares, L.L.; Macpherson, I.R.; et al. PINK1 drives production of mtDNA-containing extracellular vesicles to promote invasiveness. J. Cell Biol. 2021, 220, 6049.
- Kosaka, N.; Iguchi, H.; Hagiwara, K.; Yoshioka, Y.; Takeshita, F.; Ochiya, T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J. Biol. Chem. 2013, 288, 10849–10859.
- Zhu, J.; Lu, K.; Zhang, N.; Zhao, Y.; Ma, Q.; Shen, J.; Lin, Y.; Xiang, P.; Tang, Y.; Hu, X.; et al. Myocardial reparative functions of exosomes from mesenchymal stem cells are enhanced by hypoxia treatment of the cells via transferring microRNA-210 in an nSMase2-dependent way. Artif. Cells Nanomed. Biotechnol. 2018, 46, 1659–1670.
- Bakirtzi, K.; Man Law, I.K.; Fang, K.; Iliopoulos, D.; Pothoulakis, C. MiR-21 in Substance P-induced exosomes promotes cell proliferation and migration in human colonic epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2019, 317, G802–G810.
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659.
- Xie, S.; Zhang, Q.; Jiang, L. Current Knowledge on Exosome Biogenesis, Cargo-Sorting Mechanism and Therapeutic Implications. Membranes 2022, 12, 498.
- Fabbiano, F.; Corsi, J.; Gurrieri, E.; Trevisan, C.; Notarangelo, M.; D’Agostino, V.G. RNA packaging into extracellular vesicles: An orchestra of RNA-binding proteins? J. Extracell. Vesicles 2020, 10, e12043.
- Chen, C.; Luo, Y.; He, W.; Zhao, Y.; Kong, Y.; Liu, H.; Zhong, G.; Li, Y.; Li, J.; Huang, J.; et al. Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J. Clin. Investig. 2020, 130, 404–421.
- Chen, C.; Zheng, H.; Luo, Y.; Kong, Y.; An, M.; Li, Y.; He, W.; Gao, B.; Zhao, Y.; Huang, H.; et al. SUMOylation promotes extracellular vesicle-mediated transmission of lncRNA ELNAT1 and lymph node metastasis in bladder cancer. J. Clin. Investig. 2021, 131, 146431.
- Chen, C.; Yu, H.; Han, F.; Lai, X.; Ye, K.; Lei, S.; Mai, M.; Lai, M.; Zhang, H. Tumor-suppressive circRHOBTB3 is excreted out of cells via exosome to sustain colorectal cancer cell fitness. Mol. Cancer 2022, 21, 46.
- Pan, Z.; Zhao, R.; Li, B.; Qi, Y.; Qiu, W.; Guo, Q.; Zhang, S.; Zhao, S.; Xu, H.; Li, M.; et al. EWSR1-induced circNEIL3 promotes glioma progression and exosome-mediated macrophage immunosuppressive polarization via stabilizing IGF2BP3. Mol. Cancer 2022, 21, 16.
- Iavello, A.; Frech, V.S.; Gai, C.; Deregibus, M.C.; Quesenberry, P.J.; Camussi, G. Role of Alix in miRNA packaging during extracellular vesicle biogenesis. Int. J. Mol. Med. 2016, 37, 958–966.
- Roden, C.; Gladfelter, A.S. RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol. 2021, 22, 183–195.
- Ryan, V.H.; Dignon, G.L.; Zerze, G.H.; Chabata, C.V.; Silva, R.; Conicella, A.E.; Amaya, J.; Burke, K.A.; Mittal, J.; Fawzi, N.L. Mechanistic View of hnRNPA2 Low-Complexity Domain Structure, Interactions, and Phase Separation Altered by Mutation and Arginine Methylation. Mol. Cell 2018, 69, 465–479.e467.
- Liu, X.M.; Ma, L.; Schekman, R. Selective sorting of microRNAs into exosomes by phase-separated YBX1 condensates. eLife 2021, 10, 71982.
- Wani, S.; Kaul, D. Cancer cells govern miR-2909 exosomal recruitment through its 3’-end post-transcriptional modification. Cell Biochem. Funct. 2018, 36, 106–111.
- Yoon, T.Y.; Munson, M. SNARE complex assembly and disassembly. Curr. Biol. 2018, 28, R397–R401.
- Kou, X.; Xu, X.; Chen, C.; Sanmillan, M.L.; Cai, T.; Zhou, Y.; Giraudo, C.; Le, A.; Shi, S. The Fas/Fap-1/Cav-1 complex regulates IL-1RA secretion in mesenchymal stem cells to accelerate wound healing. Sci. Transl. Med. 2018, 10, 8524.
- Yang, L.; Peng, X.; Li, Y.; Zhang, X.; Ma, Y.; Wu, C.; Fan, Q.; Wei, S.; Li, H.; Liu, J. Long non-coding RNA HOTAIR promotes exosome secretion by regulating RAB35 and SNAP23 in hepatocellular carcinoma. Mol. Cancer 2019, 18, 78.
- Hoshino, D.; Kirkbride, K.C.; Costello, K.; Clark, E.S.; Sinha, S.; Grega-Larson, N.; Tyska, M.J.; Weaver, A.M. Exosome secretion is enhanced by invadopodia and drives invasive behavior. Cell Rep. 2013, 5, 1159–1168.
- Sinha, S.; Hoshino, D.; Hong, N.H.; Kirkbride, K.C.; Grega-Larson, N.E.; Seiki, M.; Tyska, M.J.; Weaver, A.M. Cortactin promotes exosome secretion by controlling branched actin dynamics. J. Cell Biol. 2016, 214, 197–213.
- Marat, A.L.; Ioannou, M.S.; McPherson, P.S. Connecdenn 3/DENND1C binds actin linking Rab35 activation to the actin cytoskeleton. Mol. Biol. Cell 2012, 23, 163–175.
- Beghein, E.; Devriese, D.; Van Hoey, E.; Gettemans, J. Cortactin and fascin-1 regulate extracellular vesicle release by controlling endosomal trafficking or invadopodia formation and function. Sci. Rep. 2018, 8, 15606.
- Jongsma, M.L.; Bakker, J.; Cabukusta, B.; Liv, N.; van Elsland, D.; Fermie, J.; Akkermans, J.L.; Kuijl, C.; van der Zanden, S.Y.; Janssen, L.; et al. SKIP-HOPS recruits TBC1D15 for a Rab7-to-Arl8b identity switch to control late endosome transport. EMBO J. 2020, 39, e102301.
- Granger, E.; McNee, G.; Allan, V.; Woodman, P. The role of the cytoskeleton and molecular motors in endosomal dynamics. Semin. Cell Dev. Biol. 2014, 31, 20–29.
- Jordens, I.; Fernandez-Borja, M.; Marsman, M.; Dusseljee, S.; Janssen, L.; Calafat, J.; Janssen, H.; Wubbolts, R.; Neefjes, J. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr. Biol. 2001, 11, 1680–1685.
- Wozniak, A.L.; Adams, A.; King, K.E.; Dunn, W.; Christenson, L.K.; Hung, W.T.; Weinman, S.A. The RNA binding protein FMR1 controls selective exosomal miRNA cargo loading during inflammation. J. Cell Biol. 2020, 219, 12074.
- Wei, D.; Zhan, W.; Gao, Y.; Huang, L.; Gong, R.; Wang, W.; Zhang, R.; Wu, Y.; Gao, S.; Kang, T. RAB31 marks and controls an ESCRT-independent exosome pathway. Cell Res. 2021, 31, 157–177.
- Kinchen, J.M.; Ravichandran, K.S. Identification of two evolutionarily conserved genes regulating processing of engulfed apoptotic cells. Nature 2010, 464, 778–782.
- Fei, X.; Li, Z.; Yang, D.; Kong, X.; Lu, X.; Shen, Y.; Li, X.; Xie, S.; Wang, J.; Zhao, Y.; et al. Neddylation of Coro1a determines the fate of multivesicular bodies and biogenesis of extracellular vesicles. J. Extracell. Vesicles 2021, 10, e12153.
- Villarroya-Beltri, C.; Baixauli, F.; Mittelbrunn, M.; Fernández-Delgado, I.; Torralba, D.; Moreno-Gonzalo, O.; Baldanta, S.; Enrich, C.; Guerra, S.; Sánchez-Madrid, F. ISGylation controls exosome secretion by promoting lysosomal degradation of MVB proteins. Nat. Commun. 2016, 7, 13588.
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010, 12, 19–30.
- Alnaas, A.A.; Watson-Siriboe, A.; Tran, S.; Negussie, M.; Henderson, J.A.; Osterberg, J.R.; Chon, N.L.; Harrott, B.M.; Oviedo, J.; Lyakhova, T.; et al. Multivalent lipid targeting by the calcium-independent C2A domain of synaptotagmin-like protein 4/granuphilin. J. Biol. Chem. 2021, 296, 100159.
- He, J.; Johnson, J.L.; Monfregola, J.; Ramadass, M.; Pestonjamasp, K.; Napolitano, G.; Zhang, J.; Catz, S.D. Munc13-4 interacts with syntaxin 7 and regulates late endosomal maturation, endosomal signaling, and TLR9-initiated cellular responses. Mol. Biol. Cell 2016, 27, 572–587.
- Zhang, J.; Zhang, K.; Qi, L.; Hu, Q.; Shen, Z.; Liu, B.; Deng, J.; Zhang, C.; Zhang, Y. DENN domain-containing protein FAM45A regulates the homeostasis of late/multivesicular endosomes. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 916–929.
- Imai, A.; Ishida, M.; Fukuda, M.; Nashida, T.; Shimomura, H. MADD/DENN/Rab3GEP functions as a guanine nucleotide exchange factor for Rab27 during granule exocytosis of rat parotid acinar cells. Arch. Biochem. Biophys. 2013, 536, 31–37.
- Song, L.; Tang, S.; Han, X.; Jiang, Z.; Dong, L.; Liu, C.; Liang, X.; Dong, J.; Qiu, C.; Wang, Y.; et al. KIBRA controls exosome secretion via inhibiting the proteasomal degradation of Rab27a. Nat. Commun. 2019, 10, 1639.
- Savina, A.; Vidal, M.; Colombo, M.I. The exosome pathway in K562 cells is regulated by Rab11. J. Cell Sci. 2002, 115, 2505–2515.
- Hsu, C.; Morohashi, Y.; Yoshimura, S.; Manrique-Hoyos, N.; Jung, S.; Lauterbach, M.A.; Bakhti, M.; Grønborg, M.; Möbius, W.; Rhee, J.; et al. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J. Cell Biol. 2010, 189, 223–232.
- Messenger, S.W.; Woo, S.S.; Sun, Z.; Martin, T.F.J. A Ca2+-stimulated exosome release pathway in cancer cells is regulated by Munc13-4. J. Cell Biol. 2018, 217, 2877–2890.
- Savina, A.; Fader, C.M.; Damiani, M.T.; Colombo, M.I. Rab11 promotes docking and fusion of multivesicular bodies in a calcium-dependent manner. Traffic 2005, 6, 131–143.
This entry is offline, you can click here to edit this entry!
