Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Immunology
Coronaviruses are single-stranded positive-sense RNA (+ssRNA) viruses belonging to the Coronaviridae family. Their genome encodes replicase/transcriptase proteins, structural proteins and a set of non-structural proteins linked to their virulence and proofreading activities of the replicase complex.
- SARS-CoV-2
- mRNA vaccines
- BNT162b2
- mRNA-1273
1. Introduction
Coronaviruses comprise a group of pathogens affecting many vertebrate species, from birds to humans [1]. These viruses generally cause respiratory and enteric diseases, but some coronavirus species can cause other diseases such as hepatitis or encephalitis in non-human vertebrates. Coronaviruses are single-stranded positive-sense RNA (+ssRNA) viruses belonging to the Coronaviridae family [2]. Their genome encodes replicase/transcriptase proteins, structural proteins and a set of non-structural proteins linked to their virulence and proofreading activities of the replicase complex [3,4]. The coronavirus virion contains a helical nucleoprotein made of a single +ssRNA genome bound to the nucleocapsid protein (N) [5]. This nucleoprotein is further organized into a packed internal core, enveloped by the virus membrane which derives from the endoplasmic reticulum/Golgi. The spike (S), membrane (M) and envelope (E) proteins are the main structural proteins inserted into the virus envelope [6]. The spike protein is further organized into trimers, forming the “corona” of peplomers that gives rise to the name of the family (Figure 1). In 2003, an outbreak of infectious pneumonia put the international community into high alert, leading to the discovery of severe acute respiratory syndrome coronavirus-1 (SARS-CoV-1), the first human pathogen of the family to cause lethal disease [7,8]. That first outbreak was controlled until a second one caused by a closely related coronavirus, SARS-CoV-2, originated one of the most severe pandemics in human history. The global epidemic was so dramatic that it accelerated the engineering of vaccines targeting SARS-CoV-2 at rates never seen before. The previous experience with other coronaviruses, including SARS-CoV-1 and MERS-CoV [9,10,11], prompted the selection of the spike protein as the main immunogen in most vaccines [12,13]. From the four main structural proteins, the S protein is one of the most immunogenic, at least in raising neutralizing antibody responses. Among all the potential types, mRNA vaccines soon became the primary candidates.
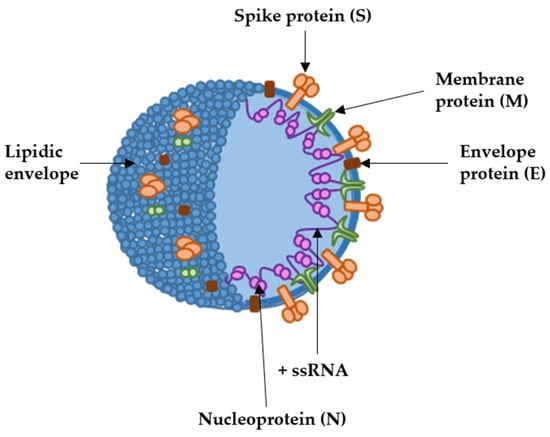
Figure 1. SARS-CoV-2 structure with the localization of the main structural proteins (arrows). The coronavirion is spherical, of about 100 to 120 nm of diameter. +ssRNA, indicates the positive-sense single-stranded RNA genome.
Vaccines based on antigen delivery through mRNA are produced by a simple and fast procedure which consists of the amplification of the RNA nucleotide sequence encoding the open reading frame of the desired gene, plus further modifications to enhance stability and translation. The procedure is performed in automatized factories in which the risk of contamination with unrelated material is very low [14].
The first mRNA vaccine for infectious diseases was developed against influenza A in 2012. In vivo experiments in mice showed specific B- and T-cell based protection. The efficacy of the vaccine was also proven in ferrets and pigs [15]. After that, various mRNA vaccines were tested in animal models to evaluate their efficacy against the Zika virus, Ebola virus, cytomegalovirus and human immunodeficiency virus (VIH), among others [16,17,18,19,20,21].
Several vaccines have been developed for SARS-CoV-2 to reduce its transmission and virulence. Among other adenoviral and protein-based vaccines, the European Medicines Agency (EMA) authorized two mRNA vaccines for human use, BNT162b2/Pfizer–BioNTech and mRNA-1273/Moderna, in December 2020 and January 2021, respectively.
The BNT162b2 vaccine consists of a lipid nanoparticle, which contains an mRNA encoding the full-length spike protein with two proline substitutions (in positions 986 and 987) in the S2 subunit to maintain the protein in the prefusion conformation [22,23]. Similarly, the mRNA-1273 vaccine consists of a lipid nanoparticle capsule constituted by four lipids, which also carries an mRNA encoding the SARS-CoV-2 full-length spike glycoprotein with the intact furin cleavage site and the two proline substitutions in the S2 subunit [24,25]. Up to three doses of these vaccines have been allowed and even four doses in vulnerable people at risk [26,27,28].
The lipid nanoparticle (LNP) of BNT162b2 is composed of ALC-0159 (2[polyethylene glycol)-2000]-N,N-ditetradecylacetamide) and DSPC (1,2-distearoyl-sn-glycero-3-phosphocholine) which play an important role in the formation of a stable lipid-bilayer nanoparticle. The LNP is structurally supported by cholesterol. Finally, the ALC-0315 ((4-hydroxybutyl) azanediyl)-bis(hexane-6,1-diyl)-bis(2-hexyldecanoate)) is the fundamental component for mRNA delivery into the cell. In addition, the vaccine is supplemented with salt buffers in order to balance the pH, and sucrose to protect the vaccine during freezing. Similarly, the mRNA-1273 LNP is stabilized by polyethylene glycol (PEG) 2000 DMG and DSPC, which form a lipid bilayer that is structurally supported by cholesterol. In contrast to BNT162b2 LNP, it carries lipid SM-102 in order to release the mRNA into the cell. This vaccine is also supplemented with salt buffers to balance the pH and with sucrose that serves as cryo-protectant.
LNP based-vaccines present a challenge due to the lack of thermostability and ultra-cold storage requirements, a fact that has limited their use in resource-poor countries. In addition, lipid and cholesterol excipients make the vaccines prone to oxidative degradation, which could decrease the stability of the vaccines. Furthermore, these BNT162b2 and mRNA-1273 mRNA vaccines, in particular, could pose a challenge in terms of delivery due to the long ribonucleic acids (4284 and 4004 nucleotides, respectively) with a number of modified nucleosides.
The BNT162b2 vaccine has been approved in 85 countries from North and South America and Europe; while mRNA-1273 has been distributed in 45 countries in Europe and North America [29].
2. Advantages and Caveats of Efficacy and Safety of mRNA SARS-CoV-2 Vaccines
The advantages of the BNT162b2 and mRNA-1273 vaccines in terms of efficacy were readily noticeable right at the beginning of their administration to the general population. The application of these vaccines was quickly associated with a decrease in COVID-19 symptomatology and spread [30]. Their fast efficacy was caused by a combination of factors: the induction of high titers of neutralizing antibodies, the activation of T-cell responses, and a demonstrated efficacy within different population groups, including vulnerable people such as the elderly.
In contrast, over the course of time, we have been aware of some of their limitations, especially after the selection and propagation of variants. The most significant are the rare but serious adverse events specifically associated with these mRNA vaccines, short-lived protection, reduced efficacy towards variants of concern and the activation of Th17 immune responses which can exacerbate inflammatory reactions.
2.1. Induction of Neutralising Antibodies and T-Cell Activation
In phase three of their respective clinical trials, vaccination with BNT162b2 and mRNA-1273 vaccines provided protection against symptomatic COVID-19 in 95 and 94.1% of the vaccinated participants, respectively. These trials were carried out in groups of subjects with ages ranging from 16 to 55. Their efficacy was also proven in older adults comprising the population most vulnerable to COVID-19 (>65 years of age) and also in adolescents (<16 years of age) [23,24,31,32,33]. Further studies confirmed the efficacy of these vaccines against the SARS-CoV-2 original strain by the fast induction of high titers of IgM and IgG antibodies specific towards the S protein, and with potent neutralizing capacities. These antibody titers remained detectable up to six months post-vaccination. Additionally, some studies evaluated the generation of T-cell responses towards S-derived peptides, demonstrating the presence of S-specific CD4 and CD8 T cells within 10 days to 9 weeks following the first and second dose, or even up to 6 months post-vaccination in healthy donors [29,34,35,36,37,38,39,40,41,42]. Furthermore, third and fourth doses led to improved immune responses compared to two doses of mRNA vaccines, leading to a peak in IgG titers in the fourth week postvaccination [43,44,45].
2.2. Efficacy in Vulnerable Populations
However, it turned out that not all vulnerable groups of people benefit from the current mRNA vaccines. This is specially the case for organ-transplanted patients or patients suffering multiple sclerosis. These patients did not benefit from BNT162b2 and mRNA-1273 vaccines due to their immunosuppressive treatments [46,47,48,49,50,51,52,53]. On the other hand, the efficacy of these vaccines has been demonstrated in patients with several types of cancers. Most studies highlight the induction of S-specific antibodies after mRNA vaccination in solid-tumor patients and oncohematological patients. This is especially true in the third week after the administration of the second dose, reaching similar numbers of antibody titers as healthy donor groups [54,55,56,57,58,59,60]. Some studies also detected CD4 and CD8 T cells specific for the S protein in solid-tumor patients up to 6 months post-vaccination. In these latter cases, the antibody titers were comparable to those achieved in healthy donors vaccinated with the mRNA vaccines [42,61,62,63]. Nevertheless, patients with hematological cancers and vaccinated with the mRNA vaccines presented decreased numbers of specific-T cells compared to healthy individuals [63,64].
2.3. Duration of Protection
As reported by several studies, S-specific IgGs induced by mRNA vaccines decrease 6 months after the second dose of mRNA vaccination [42,65]. As mentioned previously, the third and fourth dose further significantly increase IgG titers compared to titers achieved in subjects vaccinated with only two doses; however, IgG titers again decrease six months after the booster dose [44,66]. It needs to be highlighted that in patients with cancer, the persistence of antibody responses is generally shorter compared to healthy subjects following vaccination [67,68].
According to the duration of T cell responses, some studies reported the expansion of vaccine-specific T cells with a stem cell memory phenotype (TSMC). This is an important observation, because these T cells could persist for decades, providing long-term protection against SARS-CoV-2. However, in general terms, the specific CD4 and CD8 T cells are generally lost 6 months post-vaccination [69]. T-cell responses can be studied in more detail by analysing the phenotype of T cells expanded following vaccination with the mRNA vaccines. For example, CD62L and CD45RA expression in T cells was assessed by us in a recent study [42]. CD62L and CD45RA surface markers are involved in lymphocyte migration to inflammation sites and participate in T-cell receptor (TCR) signal transduction during antigen recognition [70]. In human T cells, these markers can be used to identify four types according to their differentiation degree: naïve (CD62L+ CD45RA+), central memory (CD62L+ CD45RAneg), effector memory (CD62Lneg CD45RAneg) and effector T cells (CD62Lneg CD45RAneg) [71,72]. Our study reported that both healthy individuals and patients with cancer without previous SARS-CoV-2 infection showed an expansion of effector T cells (CD62L- CD45RA+) after mRNA vaccination. However, importantly, these mRNA vaccines did not expand T cells with an effector-memory phenotype (CD62L- CD45RA-). This is in stark contrast to vaccination of individuals who had had a previous SARS-CoV-2 infection. In these subjects, vaccination achieved the expansion of effector memory T cells [42] (Figure 2).
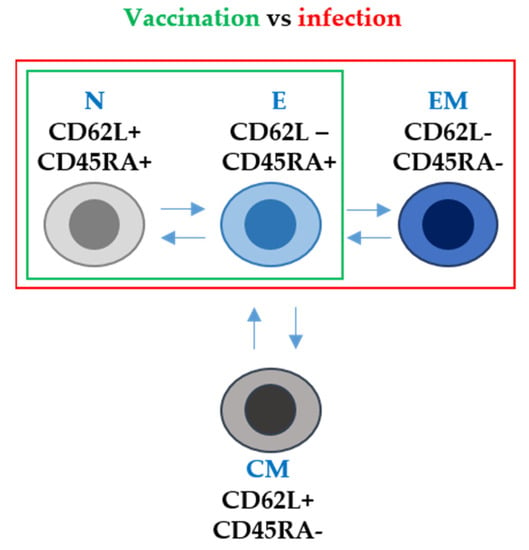
Figure 2. T-cell phenotype after mRNA vaccination and after SARS-CoV-2 infection in terms of CD62L and CD45RA surface marker expression. N, E, EM and CM stand for naïve, effector, effector memory and central memory T-cell subsets. Arrows indicate the differentiation pathways between the different T-cell phenotypes. Within the green box, T-cell subsets differentiated following mRNA vaccination only. Within the red box, T-cell subsets differentiated following infection with SARS-CoV-2.
2.4. Activation of the T-Helper 17 Responses
In many cases, SARS-CoV-2 leads to the death of the patient by exerting an exacerbated inflammatory response within the lungs of infected patients. Some studies have linked the establishment of a Th17-type of T-cell response during COVID-19 with the activation of a pro-inflammatory cytokine cascade (cytokine storm) [73,74]. For most vaccines targeting infectious agents, it would be desirable to elicit immune responses of the Th1 and Th2 types. These responses are efficacious in raising antiviral immunity while activating antibody responses. Th1 responses are regulated by T cells which mainly express IFN-gamma and IL-2, and they have a key role in attracting immune cells to the site of infection and in mediating the T-cell cytotoxicity of infected cells; Th2 responses are regulated by T cells expressing mainly IL-4 and IL-10, and they are involved in efficacious antibody production and airway inflammation observed in some respiratory diseases [75]. On the other hand, Th17 responses are regulated by T cells expressing IL-17, IL-6 and IFN-gamma. Th17 responses are fast, strong inflammatory reactions which can be critical in situations of high immunological stress. However, Th17 responses imbalance Th1-Th2 immunity, contributing to the exacerbation of inflammation, and in the case of SARS-CoV-2, its pathogenesis [73,75,76,77]. Recent studies have reported the induction of elevated concentrations of IL-17 after mRNA vaccination, indicating that mRNA vaccines trigger this strong inflammatory response [78] (Figure 3). Indeed, our study described an enhancement of this response in vaccinated oncologic patients without previous SARS-CoV-2 infection [42]. These results indicated that mRNA vaccination in patients with cancer can potentiate their chronic inflammatory status often originated and exacerbated by solid tumors, or their treatments [79,80,81,82,83,84,85].
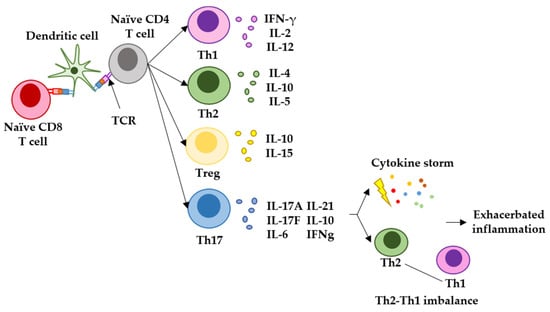
Figure 3. Representation of the main T-helper responses after antigen presentation to CD4 T cells and the role of Th17 response in generating exacerbated inflammation. Naïve CD4 and CD8 T cells recognize antigenic peptides presented by dendritic cells through the T-cell receptor (TCR). Different T-helper pathways can be activated, but the main responses observed in infectious diseases and vaccination are represented here. The Th1 response is characterized by T cells expressing IFN-gamma and IL-2, and regulate antiviral cytotoxic responses. The Th2 response is characterized by T cells expressing mainly IL-4 and IL-10, and regulated B-cell maturation to plasma cells, leading to antibody responses and airway inflammation. T-regulatory cells express mainly IL-10 and IL-15, and inhibit autoreactive damage. Finally, the Th17 response is characterized by T cells expressing mainly IL-17, IL-6, IL-21, IL-10 and IFNgamma. Th17 responses are strong inflammatory reactions caused by an immunological stress, and can lead to cytokine storms and imbalance in Th1-Th2 responses.
2.5. Loss of Efficacy towards Variants of Concern
The coronavirus S protein is the largest and most exposed antigen of the viral particle. Three molecules of the S protein form the coronavirion peplomer, which confers entry to the cell and tissue tropism [4,86]. For SARS-CoV-2, the receptor for the S protein is the ACE2 surface protein [87]. This fact makes the S gene subject to strong selective pressure from the immune system, which leads to viral escape mechanisms by increasing the number of mutations, specially concentrated in the proximities of the receptor-binding domain (RBD) (Figure 4). As most SARS-CoV-2 vaccines utilize the S protein sequence from the original Wuhan strain, these escape mutants can also escape from immune responses caused by the vaccines. This, in turn, results in a subsequent decrease in efficacy for all vaccines which use the original S protein sequence.
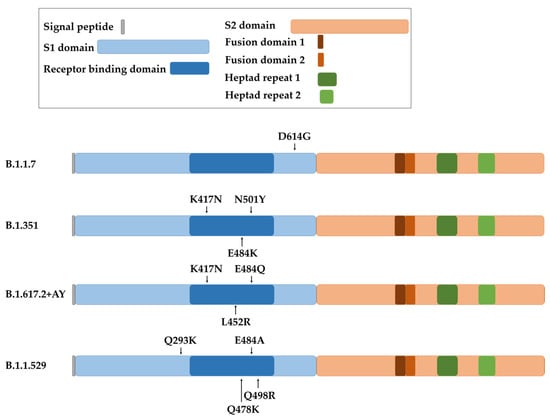
Figure 4. Representation of the spike protein of SARS-CoV-2 with the indicated domains and the localization of the main mutations involved in increased virus transmission for each variant (B.1.1.7 or Alpha variant, B.1.351 or Beta variant, B.1.617.2 + AY or Delta plus variant, and B.1.1.529 or Omicron variant).
The first dominant D614G substitution in the spike protein arose in the B.1.1.7 variant, more commonly known as Alpha SARS-CoV-2 variant (Figure 4). It has to be noted that this mutation is outside the RBD, but it nevertheless increased viral replication and transmission. Several studies later demonstrated that the D614G mutation did not decrease the protection conferred by mRNA-vaccines, which was maintained at a 94–95% of efficacy and generating comparable titers of neutralizing antibodies compared to the efficacy towards the original Wuhan strain [88,89]. Several other mutations were selected. For example, E484K, N501Y and K417N mutations in the B.1.351 variant, also known as Beta (Figure 4). These mutations were reported to cause a decline in efficacy of mRNA vaccines. The neutralizing capacities of sera from mRNA-1273- and BNT162b2-vaccinated individuals was approximately 10-fold lower towards this variant compared to the original strain [90,91]. Even so, mRNA vaccines continued to be effective against the spreading and pathogenesis of SARS-CoV-2. This was in contrast to the adenoviral-vectored ChAdOx1 vaccine, which was associated with a significant decrease in efficacy against this variant [92].
The COVID-19 strain B.1.617.2 (Delta) contained 18 novel mutations compared to the original strain [93] (Figure 4). These changes increased the transmission rate of the virus and increased its affinity to lung epithelial cells [94]. In particular, E484Q and L452R mutations enhanced immunological evasion and resistance to neutralizing antibodies from vaccinated individuals and convalescent people [95]. The protective efficacy of mRNA vaccines decreased to 88% for this variant [96], with a subsequent decrease in protection against infection six months post-vaccination [97]. In addition, the B.1.617.2 + AY sub-variants (Delta plus) selected an extra mutation (K417N) which potentiated escape from neutralizing antibodies generated by the original vaccines [94,98]. Moreover, a recent comparative study of S mutations in Alpha, Beta and Delta variants highlighted the progressive capacity of the virus strains to enter cells independently of S protein–ACE2 interactions. This fact augments transmissibility of the virus as the number of mutations increases [99]. This situation is not novel with coronaviruses, as it is likely that some coronavirus species can use a co-receptor to modulate the in vivo tissue tropism [86,100,101,102].
A variant of high interest was selected in regions with a high percentage of vaccinated population, suggesting that this variant was an escape mutant from the vaccines themselves. This variant was termed B.1.1.529, or Omicron, and its S gene accumulated more than 30 mutations compared to the original strain [98,103] (Figure 4). T478K, Q293K, Q498R and E484A contributed to an elevated transmission rate and evasion from neutralizing antibodies [104]. Due to this enhanced escaping capacity, the protection achieved with BNT162b2 and mRNA-1273 vaccines decreased to 30% after three doses, and to 47.2% in older adults [93,105,106,107,108,109,110,111,112]. This variant is still evolving, leading to Omicron sub-variants such as BQ, XBB and BF.7, with high capacities to avoid neutralizing antibodies elicited by the original vaccines [108,113].
The decline in the protection of the population against SARS-CoV-2 caused by the selection of new variants has prompted the redesign of mRNA vaccines. This is the advantage of mRNA vaccines, which allow fast modifications by just changing the immunogenic transgene to target variants. Pfizer–BioNTech and Moderna brought to the market two bivalent vaccines—Comirnaty and Spikevax—containing mRNAs encoding the spike protein of the original variant together with BA.4-5 Omicron variants [114,115]. Booster doses with these vaccines seem to offer increased protection against new Omicron subvariants, generating higher titers of Omicron-specific neutralizing antibodies than monovalent vaccines [115,116]. Nevertheless, long-term follow-up studies should be carried out to obtain more solid and robust data on the impact on the protection and spreading of the virus in the human population.
2.6. Adverse Events Caused by mRNA Vaccines
It has to be remarked that no serious adverse effects were described in the clinical trials assessing mRNA vaccines BNT162b2 and mRNA-1273 which led to their approval [22,33]. However, the administration of millions of vaccine doses has uncovered rare adverse events and complications, characterized by a diversity of symptoms. In general terms, complications from SARS-CoV-2 infection outweigh the risk of suffering these rare adverse effects following vaccination. Nevertheless, it is necessary to follow the evolution of the affected population to identify causal agents of adverse events to either improve vaccine formulations, or to better allocate the populations that need vaccination.
Two large-scale studies were carried out in the United Kingdom in about 40 million people vaccinated with sequential doses of the adenovirus-based ChAdOx1 vaccine or mRNA vaccines to evaluate cardiac adverse events. The results showed the occurrence of myocarditis in 0.004 and 0.007% of the vaccinated people with ChAdOx1 and mRNA vaccines, respectively. A statistical analysis of the data in both studies uncovered an increased risk of suffering myocarditis after the first dose of ChAdOx1 and BNT162b2 vaccine than after the further booster doses of the mRNA-1273 vaccine [117,118]. These studies indicated that especially males under 40 years of age had an elevated risk [119]. A study carried out in the USA over large-scale databases reported an elevated occurrence of myocarditis or pericarditis in mRNA-vaccinated people between 18 and 25 years of age following the second dose, without significant differences between BNT162b2 and mRNA-1273 vaccine formulations [120]. On the other hand, no clear association between vaccination and cardiac arrhythmia has been demonstrated, as most cases occurred after SARS-CoV-2 infection and this can be a confounding factor [117]. These adverse events have been therefore stated in the official websites of Comirnaty and Spikevax and by the European Medicine Agency (EMA), more specifically occurrence of myocarditis and pericarditis in some vaccinated people (https://www.comirnaty.com/, https://spikevax.com/, https://www.ema.europa.eu/en/documents/prac-recommendation/signal-assessment-report-myocarditis-pericarditis-spikevax-previously-covid-19-vaccine-moderna-covid_en.pdf, https://www.ema.europa.eu/en/documents/prac-recommendation/signal-assessment-report-myocarditis-pericarditis-spikevax-previously-covid-19-vaccine-moderna-covid_en.pdf; accessed on 20 March 2023). Isolated cases of vasospastic angina and Takotsubo cardiomyopathy have also been observed after mRNA vaccination. In addition, there are reported cases of myocardial infarction, stroke and pulmonary embolism in people older than 75 years of age after BNT162b2.
This entry is adapted from the peer-reviewed paper 10.3390/ijms24065944
This entry is offline, you can click here to edit this entry!
