Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
The mitochondrial protease called ClpP plays a central role in mitochondrial protein quality control. ClpP agonists have emerged as a novel class of mitochondria-targeting drugs. Hyperactivating ClpP induces uncontrolled, but selective, degradation of ClpP substrates and disrupts mitochondrial functions, leading to growth inhibition of breast cancer cells, without adverse effect in non-malignant cells.
- breast cancer
- mitochondria
- cancer metabolism
- ClpP
1. ClpP—The Structure and Functions
In humans, mitochondria contain over ~1000 proteins that fulfil multiple functions [1][2]. According to the latest version of MitoCarta 3.0, an inventory of mammalian mitochondrial proteins, a total of 1136 human genes, encode mitochondrial proteins [3]. Among them, 99% proteins are encoded by nuclear DNA, and 13 proteins are encoded by mtDNA. The sub-mitochondrial localizations of these proteins are the mitochondria matrix (46%), the mitochondrial inner membrane (IMM, 32%), the mitochondrial outer membrane (OMM, 10%), the mitochondrial intermembrane space (IMS, 5%), the mitochondrial membrane (3%), and unknown (5%). These proteins participate in multiple mitochondrial pathways including metabolism (40.6%), mtDNA maintenance, mtRNA metabolism and translation (20%), OxPhos (14.9%), protein import, sorting and homeostasis (7.6%), mitochondrial dynamics (9.1%), small-molecule transport (7.5%), and signaling (4.2%) [3].
Mitochondrial proteins undergo processing events, form functional assemblies, and localize to correct locations. During this process, unfolded and misfolded proteins are cleared to maintain mitochondrial protein homeostasis, which is critical for mitochondrial function [4]. Misregulation of mitochondrial proteostasis leads to human diseases, including cancers [5][6][7][8], and specialized molecular chaperones and proteases conduct this maintenance [9]. At least 45 proteases are present in the different compartments of a mitochondrion [10]. Among these proteases, the evolutionally conserved ATP-dependent proteases (AAA+ [ATPases Associated with various cellular Activities] superfamily) represent core components of the mitochondrial proteolytic system performing mitochondrial protein quality control in a mammalian cell. The members of this AAA+ proteases family are the Lon protease 1 and the ClpXP complex in the mitochondria matrix, and the i-AAA and m-AAA proteases in the inner membrane [10][11][12] (Figure 1). Among the four AAA+ proteases, ClpXP is most extensively studied in terms of its biochemical and genetic features, crystal structures, and the molecular basis of protein substrate recognition [13].
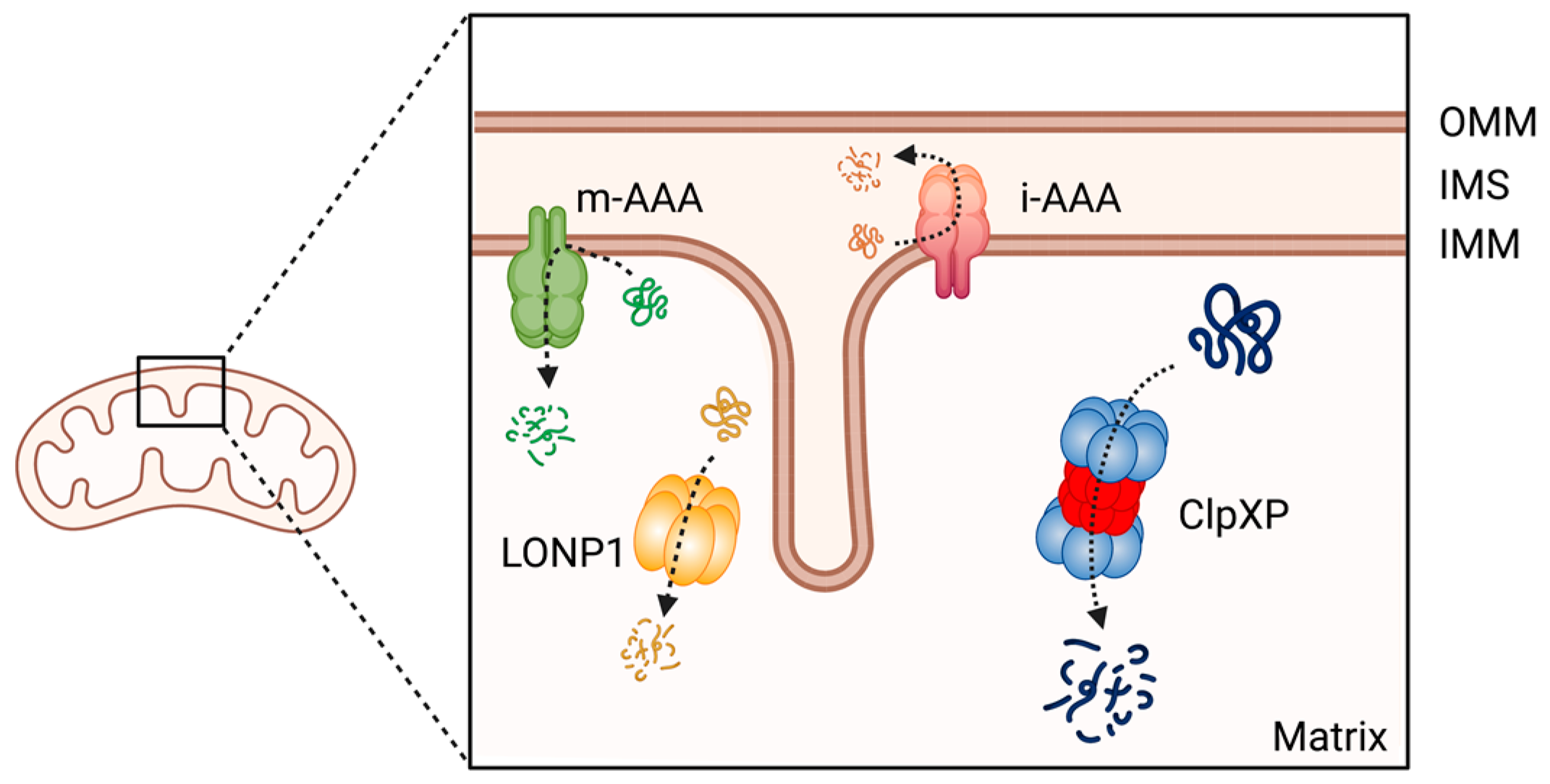
Figure 1. The member of AAA+ protease family in human mitochondria. ClpXP and LONP1 are located in the mitochondrial matrix, the i-AAA and m-AAA are located in the inner mitochondrial membrane. OMM: Outer mitochondrial membrane; IMS: intermembrane space; IMM: inner mitochondrial membrane. This figure was generated by BioRender.com (19 January 2023).
ClpXP consists of the caseinolytic mitochondrial matrix peptidase chaperone subunit X (ClpX; AAA+ ATPase) and ClpP (a tetradecameric peptidase). ClpX is a hexametric ATP-dependent protein unfoldase and translocase [14]. ClpP is a barrel-like peptidase assembled from two stacked heptameric rings, which enclose a roughly spherical proteolytic chamber [15] (Figure 2A). The proteolytic activity of ClpP is usually tightly regulated by ClpX [16]. ClpX recognizes degrons on proteins to be degraded and provides energy to unfold and translocate linearized protein into the barrel of ClpP [17]. Substrate specificity is not defined by the amino acid sequence, rather ClpX recognizes specific degradation signals and interacts with certain adaptor proteins [18]. ClpX then feeds the unfolded substrate through the axial pore to the proteolytic chamber of ClpP, and the substrate is cleaved into small peptide fragments [18][19] (Figure 2B). ClpX binds to one or both ends of ClpP and this allows for both single- and double-capped ClpXP complexes [13]. As with bacterial ClpP, human ClpP cylinders switches dynamically between extended and compact forms (Figure 2C). The active, extended form is required for substrate degradation, while an inactive compact state allows peptide product release from the ClpP inner chamber [19][20][21][22].
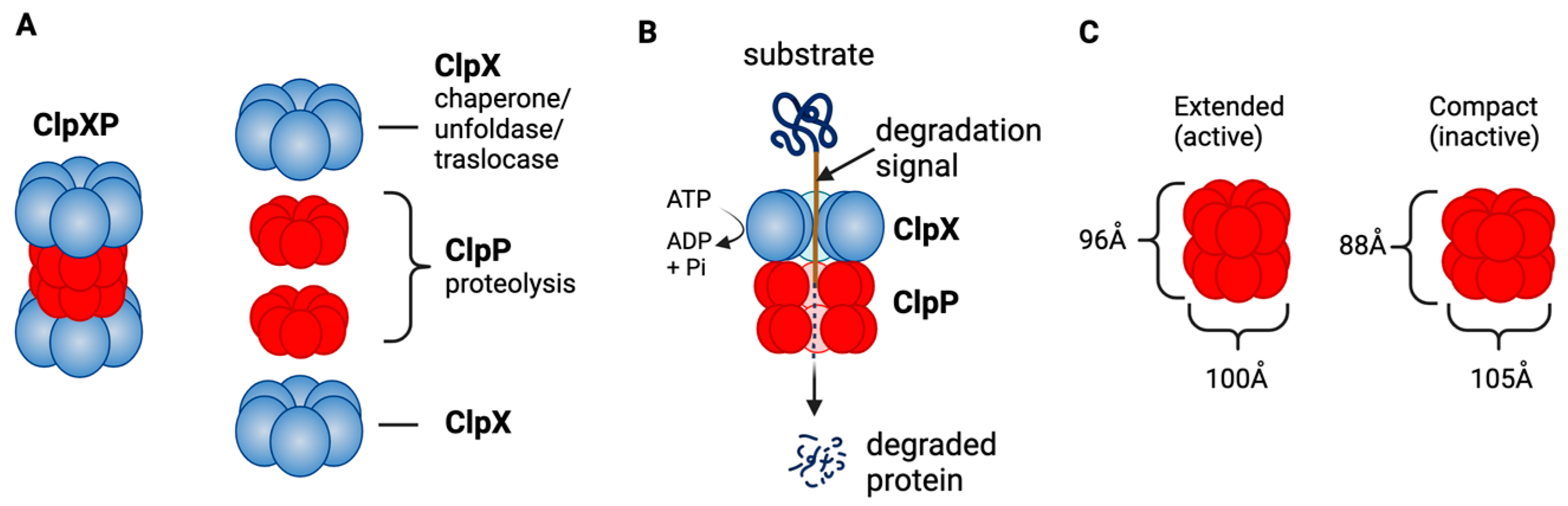
Figure 2. ClpXP structure and function as a mitochondrial protease. (A) ClpXP consists of ClpX (ATPase) and ClpP. (B) ClpX recognizes degrons of target protein, unfolds the proteins and threads them into ClpP for degradation. (C) Extended (active) and compact structure (inactive) of Staphylococcus aureus ClpP [19]. This figure was generated by BioRender.com (19 January 2023).
The role of ClpP as a master regulator of mitochondrial protein quality control and homeostasis is well established. Many mitochondrial proteins are identified as substrates of ClpXP, including proteins involved in ETC, the TCA cycle, mitochondrial ribosome, mitochondrial gene transcription and translation, glutamine metabolism, and folate metabolism [23][24][25][26]. By degrading misfolded or damaged proteins, ClpXP maintains the integrity of mitochondrial functions.
ClpP also serves as a central mediator of the mitochondrial unfolded protein response (UPRmt) [27][28], a conserved transcriptional response activated by multiple forms of mitochondrial dysfunction and regulated by mitochondrial-nuclear communication. The UPRmt supports the recovery of mitochondria from damage, restores ETC function, eliminates excess amounts of ROS, and promotes cell survival [29][30]. Recent studies indicated the prolonged UPRmt contributes to organismal deterioration including cancer [30][31][32]. In breast cancers, elevated expression of UPRmt-related genes is significantly associated with poor overall and metastasis-free survival [33]. Activated UPRmt was observed in HER2 amplified breast cancer tissues [34].
ClpP is predominantly expressed in non-malignant tissues with high mitochondrial content such as skeletal muscle, liver, and heart, suggesting its critical role in normal tissue [35]. Despite the expression in such critical organs, mice with homozygous deletion of the CLPP gene are viable, but infertile, and exhibit hearing loss and growth retardation with accumulation of ClpX and mtDNA [36]. Similarly, recessive CLPP mutations in the human CLPP genes are linked to a rare genetic disease Perrault syndrome, which shows infertility and hearing loss [37][38].
2. ClpP Activation Impairs Breast Cancer Cell Viability
While the exact roles of ClpP in tumorigenesis are not clearly established, emerging evidence demonstrate that ClpP is overexpressed in multiple malignancies including breast cancers [39][40][41]. Recent analysis of TCGA database also confirmed that ClpP is overexpressed in breast and other cancers [42]. In addition, a high level of ClpP expression is associated with the disease stage, metastasis, poor prognosis and is proposed as a prognostic marker in breast cancers [39][43][44]. Thus, ClpP is considered an emerging target for breast cancer.
Interestingly, both inhibition and hyperactivation of ClpP can lead to impaired OxPhos, resulting in cancer cell death [45]. Inhibition of ClpP induces the accumulation of damaged and misfolded mitochondrial proteins, whereas hyperactivation of ClpP leads to unregulated degradation of ClpP substrates, and both mechanisms can elicit defective cellular respiration and eventual cell death [9]. Importantly, whether inhibition or activation of ClpP induces cell death appears to be context dependent. CLPP is an essential gene for cell survival in leukemia cells and ClpP inhibition is lethal in acute myelogenous leukemia (AML), chronic myelogenous leukemia, and osteosarcoma [40]. In contrast, ClpP activators show anti-tumor effect in breast, ovary, colorectal, glioblastoma, and other cancers [23][46][47][48][49][50]. In breast cancer cells, transient knockdown of CLPP by siRNA induced apoptosis and inhibited cell viability, migration, invasion in breast cancer cells [43], while CRISPR/Cas9-mediated deletion of CLPP gene did not affect cell viability presumably due to cellular adaptation [23][48]. In contrast, activation of ClpP by ClpP agonists exerts an anti-tumor effect in breast cancer cells [23][48]. This suggests that ClpP activation, but not ClpP inhibition, is the better targeting approach in breast cancers.
3. ClpP Activating Drugs
Since ClpP activators, but not ClpP inhibitors, show cytotoxicity in breast cancers, the research will focus on ClpP activators in this entry (for ClpP inhibitors, the researchers refer readers to other excellent reviews [9][42][44]. There are three classes of ClpP agonists: acyldepsipeptide antibiotics (ADEPs), ONC201 and its analogs (imipridones), and TR compounds. The representative ClpP agonists and structures are illustrated in Figure 3.
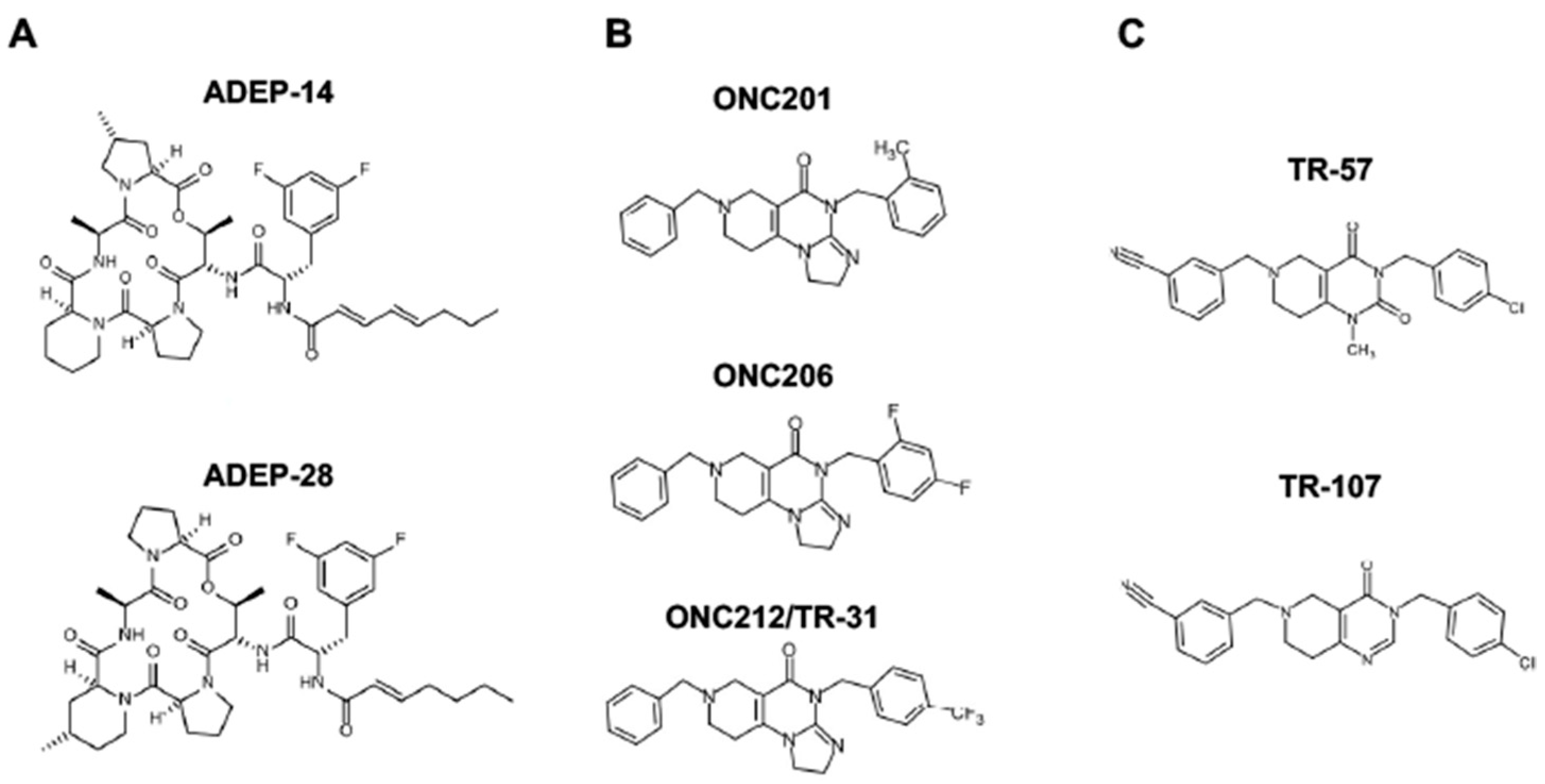
Figure 3. Representative ClpP agonists and structures. (A) Acyldepsipeptide antibiotics (ADEPs), (B) ONC201 and its analogs (imipridones), and (C) TR compounds.
ADEPs: ClpP has emerged as an anti-bacterial target during the mechanism of action studies on acyldepsipeptide antibiotics (ADEPs) in last two decades. ADEPs were first reported as antibiotics [51]. Identification of the resistance-mediating mutation within an ADEP-resistant Escherichia coli mutant and affinity chromatography with an immobilized ADEP analog led to identification of ClpP as the direct target of ADEPs [52]. ADEPs increase the activity of the bacterial ClpP, leading to bacterial cell death. As ClpP plays important roles in the bacterial virulence due to the broad range of substrates, extensive effort has been made to develop ADEPs as novel therapeutics for drug-resistant bacteria [18][52][53][54][55]. Medicinal chemistry studies revealed the structure–activity relationship and yielded many derivatives with enhanced in vitro potency and stability [56][57][58][59]. The anti-bacterial efficacy of ADEPs has been demonstrated in lethal bacterial infections in rodent models [52][56][60]. Wong et al. were the first to demonstrate that ADEPs binds to human ClpP (HsClpP) using crystallographic structural analysis [61]. ADEPs induce caspase-dependent apoptosis, mitochondrial fragmentation, and OxPhos inhibition in HEK293 cells. At present, ADEP analogues are being tested in preclinical models in breast cancers, but the findings are limited.
ONC201 and its analogs (Imipridones): ONC201 (a.k.a., TIC10) is the first-in-class small molecule of the imipridone family [62]. ONC201 was identified in a chemical library screen to find a small molecule that transcriptionally induces tumor necrosis factor-alpha related apoptosis-inducing ligand (TRAIL), leading to an autocrine induction of apoptosis in the cancer cell. The proposed mechanism of action of ONC201 was that it inhibits extracellular signal-regulated kinase (ERK) and AKT, leading to the translocation of Foxo3a into the nucleus, where it activates transcription of the TRAIL gene [62]. Subsequent studies evaluated ONC201 as an antagonist of dopamine receptors (DRD)2/3 [63][64][65]. There were some preclinical data supportive of DRD2 antagonistic activity [64], and the clinical activity seen in diffuse intrinsic pontine gliomas (DIPG) has been suggested to be mediated through DRD2 inhibition [66][67]. However, this remains controversial. No direct evidence of ONC201 binding to DRD2/3 has been shown, gene suppression or deletion of DRD2/3 did not abrogate the ONC201’s cytotoxic effect [64], and DRD2 transcript is not detectable in many breast cancer cell lines that are sensitive to ONC201 [47]. This requires further evaluation to determine if any activity of ONC201 or other ClpP agonists is due to DRD2 antagonism.
While ONC201 was shown to increase Poly (ADP-ribose) polymerase 1 (PARP) cleavage and apoptosis in some cancer models [62][68], apoptosis was not observed in breast cancers [47][48]. The researcher's group was the first to report that the cytotoxicity of ONC201 is due to targeting mitochondria and is independent of TRAIL-mediated apoptosis [47]. The researchers found that ONC201 inhibits OxPhos, depletes ATP, mtDNA, multiple mitochondrial proteins, such as mitochondrial transcription factor A (TFAM), ETC proteins, accompanied with mitochondrial structural damage and an integrated stress response (ISR) indicated by ATF4 and CHOP induction [47]. The researchers also found that breast cancer cells that lack functional mitochondria (e.g., rho0 cells) were ONC201-resistant. The researcher's finding was confirmed and extended by two studies which demonstrated that ONC201 binds to and activates ClpP using crystallography structural analysis, mass-spectrometry, and affinity chromatography/drug competition assays [46][49]. Another group identified ClpP as the target of ONC201 using a genome wide CRISPR KO library screen [69]. Later, ONC206 and ONC212 were developed by Chimerix, Inc. (https://www.chimerix.com/, accessed on 2 January 2023) as more potent imipridones compared with ONC201. Studies confirmed that these imipridones impair OxPhos, induce mitochondrial damage, ROS, and ISR [49][70]. ONC201 has been shown to penetrate the brain-blood barrier [62].
TR compounds: TR compounds are a novel series of imipridone-derived compounds and related chemicals being developed by Madera Therapeutics, LLC (https://maderat.com/, accessed on 2 January 2023). Multiple TR compounds, including TR-57, TR-31 (=ONC212), were found to be ∼50–100-fold more potent compared with ONC201 in cytotoxicity in breast cancer cells. TR compounds induced an ISR at nanomolar concentrations, significantly lower compared with those induced by ONC201 at micromolar concentrations [46]. Affinity chromatography/drug competition assays demonstrated that the TR compounds bind to ClpP with ∼10-fold higher affinity compared to ONC201 [46]. Similar to ONC201, TR compounds reduce target protein levels (e.g., TFAM) and impair OxPhos [23][46][47].
The unique feature of all ClpP agonists described above is that they activate ClpP in the absence of ClpX (Figure 4 and Figure 5). The X-ray crystallography structure demonstrated that ADEP binds to the hydrophobic pockets of ClpP and dissociates the ClpXP complex at substoichiometric concentrations, leading to ClpP activation independent of regulatory subunit ClpX [61] (Figure 4A). Similar to ADEPs, imipridones directly binds to ClpP, and displace ClpX, and the bindings triggers the opening of channel-like pore of ClpP, and thereby increase its protease activity without the ClpX [9][49][71] (Figure 4B). Recently, X-ray crystallography demonstrated that TR compounds bind to ClpP, with enhanced binding affinities due to their greater shape and charge complementarity with the surface hydrophobic pockets of ClpP [72] (Figure 5C). Moreover, N-terminome profiling of MDA-MB-231 cell line upon treatment with one of TR compounds revealed the global proteomic changes and characterized the sequence and structural properties for protein cleavage generated upon TR compound-induced ClpP-dependent proteolysis of cellular proteins [72].
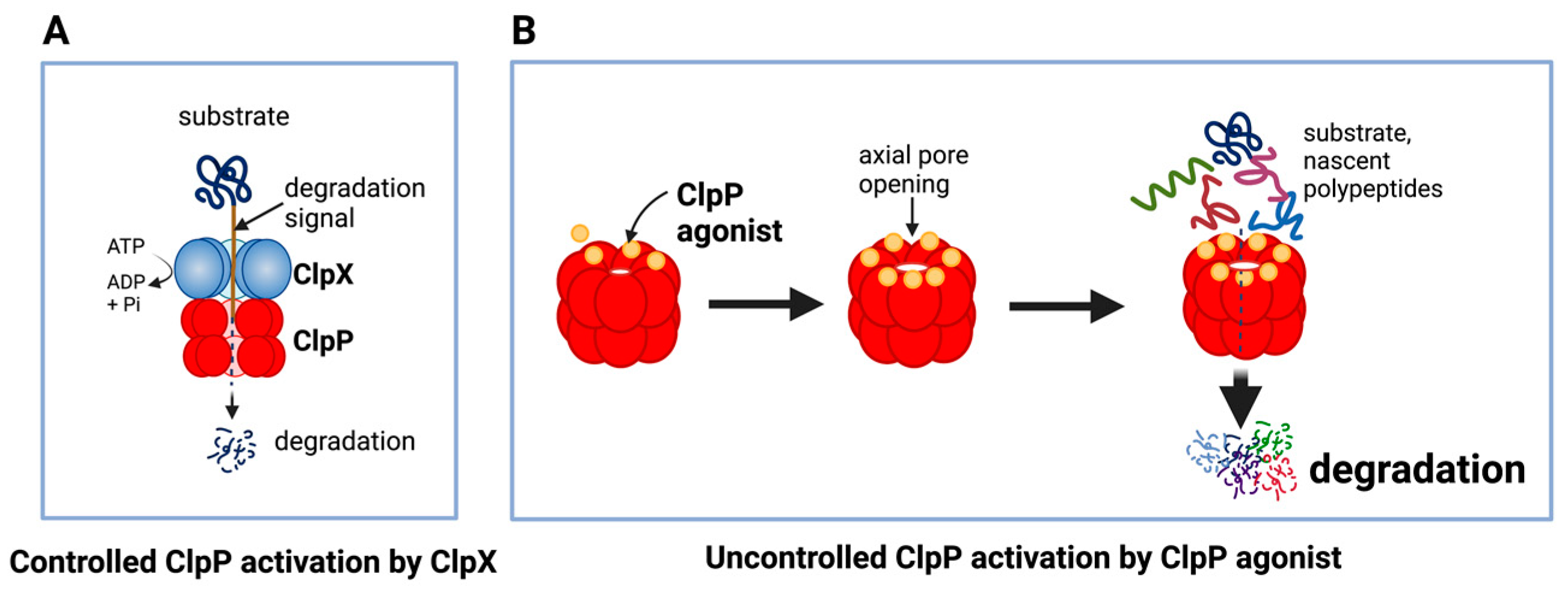
Figure 4. Controlled and uncontrolled proteolysis by ClpP. (A) In controlled proteolysis, target substrates are specifically recognized and unfolded by ClpX. The unfolded substrate is transferred into the barrel-like chamber of the ClpP, and proteolysis is carried out by active proteases at the inner surface of the chamber. (B) Uncontrolled proteolysis triggered by binding of ClpP agonists to ClpP. Binding of ClpP agonists (yellow spheres) to ClpP subunits displaces ClpX and triggers an opening of the entrance pore of the ClpP barrel, leading to unregulated degradation of nascent polypeptides and unfolded proteins. This uncontrolled ClpP activation does not require ClpX. This figure was generated by BioRender.com (20 January 2023).
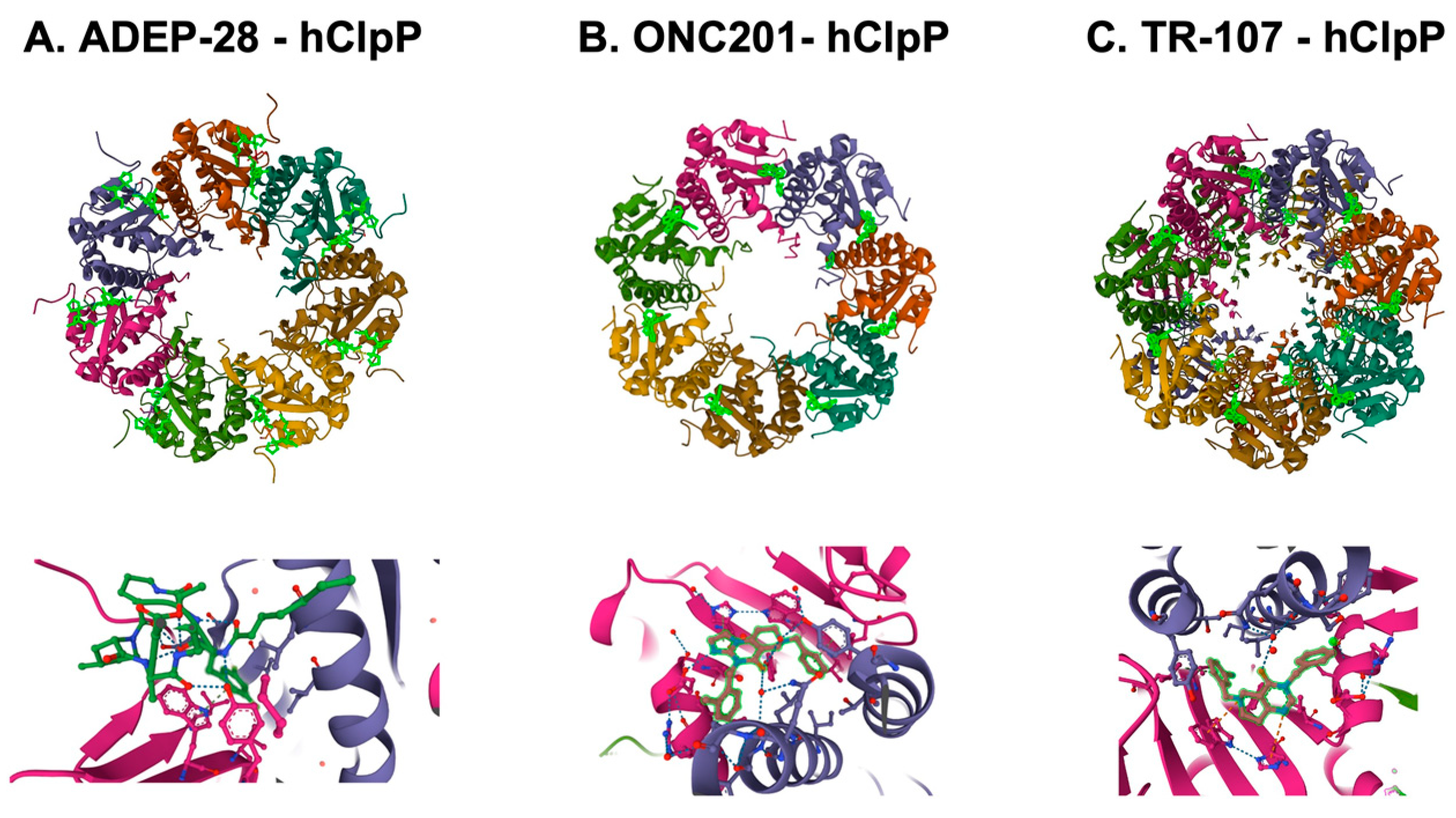
Figure 5. Crystal structures of human mitochondrial ClpP complex with ClpP ligands. (A) ADEP-28-hClpP binding [61] (PDB DOI: 10.2210/pdb6BBA/pdb), (B) ONC201-hClpP [49] (PDB DOI: 10.2210/pdb6DL7/pdb), (C) TR-107-hClpP [72]. (PDB DOI: 10.2210/pdb7UVU/pdb). Top panels show that each ClpP ligand (bright green) bound to ClpP. One heptamer ring for ADEP-28 and ONC201, or two heptamer rings for TR-107 are shown. Each subunit of ClpP is shown in a different color. Bottom panels show a magnified view of ligands (bright green) bound to the pocket between two ClpP subunits.
This entry is adapted from the peer-reviewed paper 10.3390/cancers15071936
References
- Pagliarini, D.J.; Calvo, S.E.; Chang, B.; Sheth, S.A.; Vafai, S.B.; Ong, S.E.; Walford, G.A.; Sugiana, C.; Boneh, A.; Chen, W.K.; et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell 2008, 134, 112–123.
- Morgenstern, M.; Stiller, S.B.; Lübbert, P.; Peikert, C.D.; Dannenmaier, S.; Drepper, F.; Weill, U.; Höß, P.; Feuerstein, R.; Gebert, M.; et al. Definition of a High-Confidence Mitochondrial Proteome at Quantitative Scale. Cell Rep. 2017, 19, 2836–2852.
- Rath, S.; Sharma, R.; Gupta, R.; Ast, T.; Chan, C.; Durham, T.J.; Goodman, R.P.; Grabarek, Z.; Haas, M.E.; Hung, W.H.W.; et al. MitoCarta3.0: An updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 2021, 49, D1541–D1547.
- Varabyova, A.; Stojanovski, D.; Chacinska, A. Mitochondrial protein homeostasis. IUBMB Life 2013, 65, 191–201.
- Levytskyy, R.M.; Germany, E.M.; Khalimonchuk, O. Mitochondrial Quality Control Proteases in Neuronal Welfare. J. Neuroimmune Pharmacol. 2016, 11, 629–644.
- Bulteau, A.L.; Bayot, A. Mitochondrial proteases and cancer. Biochim. Biophys. Acta 2011, 1807, 595–601.
- Rugarli, E.I.; Langer, T. Mitochondrial quality control: A matter of life and death for neurons. EMBO J. 2012, 31, 1336–1349.
- König, T.; Tröder, S.E.; Bakka, K.; Korwitz, A.; Richter-Dennerlein, R.; Lampe, P.A.; Patron, M.; Mühlmeister, M.; Guerrero-Castillo, S.; Brandt, U.; et al. The m-AAA Protease Associated with Neurodegeneration Limits MCU Activity in Mitochondria. Mol. Cell 2016, 64, 148–162.
- Nouri, K.; Feng, Y.; Schimmer, A.D. Mitochondrial ClpP serine protease-biological function and emerging target for cancer therapy. Cell Death Dis. 2020, 11, 841.
- Deshwal, S.; Fiedler, K.U.; Langer, T. Mitochondrial Proteases: Multifaceted Regulators of Mitochondrial Plasticity. Annu. Rev. Biochem. 2020, 89, 501–528.
- Song, J.; Herrmann, J.M.; Becker, T. Quality control of the mitochondrial proteome. Nat. Rev. Mol. Cell Biol. 2021, 22, 54–70.
- Ng, M.Y.W.; Wai, T.; Simonsen, A. Quality control of the mitochondrion. Dev. Cell 2021, 56, 881–905.
- Sauer, R.T.; Fei, X.; Bell, T.A.; Baker, T.A. Structure and function of ClpXP, a AAA+ proteolytic machine powered by probabilistic ATP hydrolysis. Crit. Rev. Biochem. Mol. Biol. 2022, 57, 188–204.
- Glynn, S.E.; Martin, A.; Nager, A.R.; Baker, T.A.; Sauer, R.T. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell 2009, 139, 744–756.
- Wang, J.; Hartling, J.A.; Flanagan, J.M. The structure of ClpP at 2.3 A resolution suggests a model for ATP-dependent proteolysis. Cell 1997, 91, 447–456.
- Lee, M.E.; Baker, T.A.; Sauer, R.T. Control of substrate gating and translocation into ClpP by channel residues and ClpX binding. J. Mol. Biol. 2010, 399, 707–718.
- Baker, T.A.; Sauer, R.T. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim. Biophys. Acta 2012, 1823, 15–28.
- Malik, I.T.; Brötz-Oesterhelt, H. Conformational control of the bacterial Clp protease by natural product antibiotics. Nat. Prod. Rep. 2017, 34, 815–831.
- Liu, K.; Ologbenla, A.; Houry, W.A. Dynamics of the ClpP serine protease: A model for self-compartmentalized proteases. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 400–412.
- Stahl, M.; Sieber, S.A. An amino acid domino effect orchestrates ClpP’s conformational states. Curr. Opin. Chem. Biol. 2017, 40, 102–110.
- Gribun, A.; Kimber, M.S.; Ching, R.; Sprangers, R.; Fiebig, K.M.; Houry, W.A. The ClpP double ring tetradecameric protease exhibits plastic ring-ring interactions, and the N termini of its subunits form flexible loops that are essential for ClpXP and ClpAP complex formation. J. Biol. Chem. 2005, 280, 16185–16196.
- Lowth, B.R.; Kirstein-Miles, J.; Saiyed, T.; Brötz-Oesterhelt, H.; Morimoto, R.I.; Truscott, K.N.; Dougan, D.A. Substrate recognition and processing by a Walker B mutant of the human mitochondrial AAA+ protein CLPX. J. Struct. Biol. 2012, 179, 193–201.
- Greer, Y.E.; Hernandez, L.; Fennell, E.M.J.; Kundu, M.; Voeller, D.; Chari, R.; Gilbert, S.F.; Gilbert, T.S.K.; Ratnayake, S.; Tang, B.; et al. Mitochondrial Matrix Protease ClpP Agonists Inhibit Cancer Stem Cell Function in Breast Cancer Cells by Disrupting Mitochondrial Homeostasis. Cancer Res. Commun. 2022, 2, 1144–1161.
- Mabanglo, M.F.; Bhandari, V.; Houry, W.A. Substrates and interactors of the ClpP protease in the mitochondria. Curr. Opin. Chem. Biol. 2021, 66, 102078.
- Fischer, F.; Langer, J.D.; Osiewacz, H.D. Identification of potential mitochondrial CLPXP protease interactors and substrates suggests its central role in energy metabolism. Sci. Rep. 2015, 5, 18375.
- Nguyen, T.A.; Gronauer, T.F.; Nast-Kolb, T.; Sieber, S.A.; Lang, K. Substrate Profiling of Mitochondrial Caseinolytic Protease P via a Site-Specific Photocrosslinking Approach. Angew. Chem. Int. Ed. Engl. 2022, 61, e202111085.
- Haynes, C.M.; Petrova, K.; Benedetti, C.; Yang, Y.; Ron, D. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev. Cell 2007, 13, 467–480.
- Wu, G.; Xiong, Q.; Wei, X.; Wang, Y.; Hu, X.; He, G.; Liu, L.; Lai, Q.; Dai, Z.; Anushesh, D.; et al. Mitochondrial unfolded protein response gene CLPP changes mitochondrial dynamics and affects mitochondrial function. PeerJ 2019, 7, e7209.
- Wang, G.; Fan, Y.; Cao, P.; Tan, K. Insight into the mitochondrial unfolded protein response and cancer: Opportunities and challenges. Cell Biosci. 2022, 12, 18.
- Shpilka, T.; Haynes, C.M. The mitochondrial UPR: Mechanisms, physiological functions and implications in ageing. Nat. Rev. Mol. Cell Biol. 2018, 19, 109–120.
- Inigo, J.R.; Chandra, D. The mitochondrial unfolded protein response (UPR). J. Hematol. Oncol. 2022, 15, 98.
- Inigo, J.R.; Kumar, R.; Chandra, D. Targeting the mitochondrial unfolded protein response in cancer: Opportunities and challenges. Trends Cancer 2021, 7, 1050–1053.
- Kenny, T.C.; Craig, A.J.; Villanueva, A.; Germain, D. Mitohormesis Primes Tumor Invasion and Metastasis. Cell Rep. 2019, 27, 2292–2303.e2296.
- Chen, F.M.; Huang, L.J.; Ou-Yang, F.; Kan, J.Y.; Kao, L.C.; Hou, M.F. Activation of mitochondrial unfolded protein response is associated with Her2-overexpression breast cancer. Breast Cancer Res. Treat. 2020, 183, 61–70.
- Bross, P.; Andresen, B.S.; Knudsen, I.; Kruse, T.A.; Gregersen, N. Human ClpP protease: cDNA sequence, tissue-specific expression and chromosomal assignment of the gene. FEBS Lett. 1995, 377, 249–252.
- Gispert, S.; Parganlija, D.; Klinkenberg, M.; Dröse, S.; Wittig, I.; Mittelbronn, M.; Grzmil, P.; Koob, S.; Hamann, A.; Walter, M.; et al. Loss of mitochondrial peptidase Clpp leads to infertility, hearing loss plus growth retardation via accumulation of CLPX, mtDNA and inflammatory factors. Hum. Mol. Genet. 2013, 22, 4871–4887.
- Jenkinson, E.M.; Rehman, A.U.; Walsh, T.; Clayton-Smith, J.; Lee, K.; Morell, R.J.; Drummond, M.C.; Khan, S.N.; Naeem, M.A.; Rauf, B.; et al. Perrault syndrome is caused by recessive mutations in CLPP, encoding a mitochondrial ATP-dependent chambered protease. Am. J. Hum. Genet. 2013, 92, 605–613.
- Brodie, E.J.; Zhan, H.; Saiyed, T.; Truscott, K.N.; Dougan, D.A. Perrault syndrome type 3 caused by diverse molecular defects in CLPP. Sci. Rep. 2018, 8, 12862.
- Seo, J.H.; Rivadeneira, D.B.; Caino, M.C.; Chae, Y.C.; Speicher, D.W.; Tang, H.Y.; Vaira, V.; Bosari, S.; Palleschi, A.; Rampini, P.; et al. The Mitochondrial Unfoldase-Peptidase Complex ClpXP Controls Bioenergetics Stress and Metastasis. PLoS Biol. 2016, 14, e1002507.
- Cole, A.; Wang, Z.; Coyaud, E.; Voisin, V.; Gronda, M.; Jitkova, Y.; Mattson, R.; Hurren, R.; Babovic, S.; Maclean, N.; et al. Inhibition of the Mitochondrial Protease ClpP as a Therapeutic Strategy for Human Acute Myeloid Leukemia. Cancer Cell 2015, 27, 864–876.
- Cormio, A.; Musicco, C.; Gasparre, G.; Cormio, G.; Pesce, V.; Sardanelli, A.M.; Gadaleta, M.N. Increase in proteins involved in mitochondrial fission, mitophagy, proteolysis and antioxidant response in type I endometrial cancer as an adaptive response to respiratory complex I deficiency. Biochem. Biophys. Res. Commun. 2017, 491, 85–90.
- Zhang, J.; Qiao, W.; Luo, Y. Mitochondrial quality control proteases and their modulation for cancer therapy. Med. Res. Rev. 2022, 43, 399–436.
- Luo, J.; Zeng, B.; Tao, C.; Lu, M.; Ren, G. ClpP regulates breast cancer cell proliferation, invasion and apoptosis by modulating the Src/PI3K/Akt signaling pathway. PeerJ 2020, 8, e8754.
- Cormio, A.; Sanguedolce, F.; Pesce, V.; Musicco, C. Mitochondrial Caseinolytic Protease P: A Possible Novel Prognostic Marker and Therapeutic Target in Cancer. Int. J. Mol. Sci. 2021, 22, 6228.
- Wong, K.S.; Houry, W.A. Chemical Modulation of Human Mitochondrial ClpP: Potential Application in Cancer Therapeutics. ACS Chem. Biol. 2019, 14, 2349–2360.
- Graves, P.R.; Aponte-Collazo, L.J.; Fennell, E.M.J.; Graves, A.C.; Hale, A.E.; Dicheva, N.; Herring, L.E.; Gilbert, T.S.K.; East, M.P.; McDonald, I.M.; et al. Mitochondrial Protease ClpP is a Target for the Anticancer Compounds ONC201 and Related Analogues. ACS Chem. Biol. 2019, 14, 1020–1029.
- Greer, Y.E.; Porat-Shliom, N.; Nagashima, K.; Stuelten, C.; Crooks, D.; Koparde, V.N.; Gilbert, S.F.; Islam, C.; Ubaldini, A.; Ji, Y.; et al. ONC201 kills breast cancer cells in vitro by targeting mitochondria. Oncotarget 2018, 9, 18454–18479.
- Fennell, E.M.J.; Aponte-Collazo, L.J.; Wynn, J.D.; Drizyte-Miller, K.; Leung, E.; Greer, Y.E.; Graves, P.R.; Iwanowicz, A.A.; Ashamalla, H.; Holmuhamedov, E.; et al. Characterization of TR-107, a novel chemical activator of the human mitochondrial protease ClpP. Pharmacol. Res. Perspect. 2022, 10, e00993.
- Ishizawa, J.; Zarabi, S.F.; Davis, R.E.; Halgas, O.; Nii, T.; Jitkova, Y.; Zhao, R.; St-Germain, J.; Heese, L.E.; Egan, G.; et al. Mitochondrial ClpP-Mediated Proteolysis Induces Selective Cancer Cell Lethality. Cancer Cell 2019, 35, 721–737.e729.
- Nguyen, T.T.T.; Shang, E.; Schiffgens, S.; Torrini, C.; Shu, C.; Akman, H.O.; Prabhu, V.V.; Allen, J.E.; Westhoff, M.A.; Karpel-Massler, G.; et al. Induction of Synthetic Lethality by Activation of Mitochondrial ClpP and Inhibition of HDAC1/2 in Glioblastoma. Clin. Cancer Res. 2022, 28, 1881–1895.
- Bhandary, L.; Bailey, P.C.; Chang, K.T.; Underwood, K.F.; Lee, C.J.; Whipple, R.A.; Jewell, C.M.; Ory, E.; Thompson, K.N.; Ju, J.A.; et al. Lipid tethering of breast tumor cells reduces cell aggregation during mammosphere formation. Sci. Rep. 2021, 11, 3214.
- Brötz-Oesterhelt, H.; Beyer, D.; Kroll, H.P.; Endermann, R.; Ladel, C.; Schroeder, W.; Hinzen, B.; Raddatz, S.; Paulsen, H.; Henninger, K.; et al. Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat. Med. 2005, 11, 1082–1087.
- Moreno-Cinos, C.; Goossens, K.; Salado, I.G.; Van Der Veken, P.; De Winter, H.; Augustyns, K. ClpP Protease, a Promising Antimicrobial Target. Int. J. Mol. Sci. 2019, 20, 2232.
- Leung, E.; Datti, A.; Cossette, M.; Goodreid, J.; McCaw, S.E.; Mah, M.; Nakhamchik, A.; Ogata, K.; El Bakkouri, M.; Cheng, Y.Q.; et al. Activators of cylindrical proteases as antimicrobials: Identification and development of small molecule activators of ClpP protease. Chem. Biol. 2011, 18, 1167–1178.
- Binepal, G.; Mabanglo, M.F.; Goodreid, J.D.; Leung, E.; Barghash, M.M.; Wong, K.S.; Lin, F.; Cossette, M.; Bansagi, J.; Song, B.; et al. Development of Antibiotics That Dysregulate the. ACS Infect. Dis. 2020, 6, 3224–3236.
- Conlon, B.P.; Nakayasu, E.S.; Fleck, L.E.; LaFleur, M.D.; Isabella, V.M.; Coleman, K.; Leonard, S.N.; Smith, R.D.; Adkins, J.N.; Lewis, K. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 2013, 503, 365–370.
- Socha, A.M.; Tan, N.Y.; LaPlante, K.L.; Sello, J.K. Diversity-oriented synthesis of cyclic acyldepsipeptides leads to the discovery of a potent antibacterial agent. Bioorg. Med. Chem. 2010, 18, 7193–7202.
- Carney, D.W.; Schmitz, K.R.; Truong, J.V.; Sauer, R.T.; Sello, J.K. Restriction of the conformational dynamics of the cyclic acyldepsipeptide antibiotics improves their antibacterial activity. J. Am. Chem. Soc. 2014, 136, 1922–1929.
- Goodreid, J.D.; Janetzko, J.; Santa Maria, J.P.; Wong, K.S.; Leung, E.; Eger, B.T.; Bryson, S.; Pai, E.F.; Gray-Owen, S.D.; Walker, S.; et al. Development and Characterization of Potent Cyclic Acyldepsipeptide Analogues with Increased Antimicrobial Activity. J. Med. Chem. 2016, 59, 624–646.
- Hinzen, B.; Raddatz, S.; Paulsen, H.; Lampe, T.; Schumacher, A.; Häbich, D.; Hellwig, V.; Benet-Buchholz, J.; Endermann, R.; Labischinski, H.; et al. Medicinal chemistry optimization of acyldepsipeptides of the enopeptin class antibiotics. ChemMedChem 2006, 1, 689–693.
- Wong, K.S.; Mabanglo, M.F.; Seraphim, T.V.; Mollica, A.; Mao, Y.Q.; Rizzolo, K.; Leung, E.; Moutaoufik, M.T.; Hoell, L.; Phanse, S.; et al. Acyldepsipeptide Analogs Dysregulate Human Mitochondrial ClpP Protease Activity and Cause Apoptotic Cell Death. Cell Chem. Biol. 2018, 25, 1017–1030.e1019.
- Allen, J.E.; Krigsfeld, G.; Mayes, P.A.; Patel, L.; Dicker, D.T.; Patel, A.S.; Dolloff, N.G.; Messaris, E.; Scata, K.A.; Wang, W.; et al. Dual inactivation of Akt and ERK by TIC10 signals Foxo3a nuclear translocation, TRAIL gene induction, and potent antitumor effects. Sci. Transl. Med. 2013, 5, 171ra117.
- Stein, M.N.; Bertino, J.R.; Kaufman, H.L.; Mayer, T.; Moss, R.; Silk, A.; Chan, N.; Malhotra, J.; Rodriguez, L.; Aisner, J.; et al. First-in-Human Clinical Trial of Oral ONC201 in Patients with Refractory Solid Tumors. Clin. Cancer Res. 2017, 23, 4163–4169.
- Kline, C.L.B.; Ralff, M.D.; Lulla, A.R.; Wagner, J.M.; Abbosh, P.H.; Dicker, D.T.; Allen, J.E.; El-Deiry, W.S. Role of Dopamine Receptors in the Anticancer Activity of ONC201. Neoplasia 2018, 20, 80–91.
- Prabhu, V.V.; Morrow, S.; Rahman Kawakibi, A.; Zhou, L.; Ralff, M.; Ray, J.; Jhaveri, A.; Ferrarini, I.; Lee, Y.; Parker, C.; et al. ONC201 and imipridones: Anti-cancer compounds with clinical efficacy. Neoplasia 2020, 22, 725–744.
- Chi, A.S.; Tarapore, R.S.; Hall, M.D.; Shonka, N.; Gardner, S.; Umemura, Y.; Sumrall, A.; Khatib, Z.; Mueller, S.; Kline, C.; et al. Pediatric and adult H3 K27M-mutant diffuse midline glioma treated with the selective DRD2 antagonist ONC201. J. Neurooncol. 2019, 145, 97–105.
- Hall, M.D.; Odia, Y.; Allen, J.E.; Tarapore, R.; Khatib, Z.; Niazi, T.N.; Daghistani, D.; Schalop, L.; Chi, A.S.; Oster, W.; et al. First clinical experience with DRD2/3 antagonist ONC201 in H3 K27M-mutant pediatric diffuse intrinsic pontine glioma: A case report. J. Neurosurg. Pediatr. 2019, 23, 719–725.
- Ni, X.; Zhang, X.; Hu, C.H.; Langridge, T.; Tarapore, R.S.; Allen, J.E.; Oster, W.; Duvic, M. ONC201 selectively induces apoptosis in cutaneous T-cell lymphoma cells via activating pro-apoptotic integrated stress response and inactivating JAK/STAT and NF-κB pathways. Oncotarget 2017, 8, 61761–61776.
- Jacques, S.; van der Sloot, A.M.; C Huard, C.; Coulombe-Huntington, J.; Tsao, S.; Tollis, S.; Bertomeu, T.; Culp, E.J.; Pallant, D.; Cook, M.A.; et al. Imipridone Anticancer Compounds Ectopically Activate the ClpP Protease and Represent a New Scaffold for Antibiotic Development. Genetics 2020, 214, 1103–1120.
- Przystal, J.M.; Cianciolo Cosentino, C.; Yadavilli, S.; Zhang, J.; Laternser, S.; Bonner, E.R.; Prasad, R.; Dawood, A.A.; Lobeto, N.; Chin Chong, W.; et al. Imipridones affect tumor bioenergetics and promote cell lineage differentiation in diffuse midline gliomas. Neuro. Oncol. 2022, 24, 1438–1451.
- Mabanglo, M.F.; Houry, W.A. Recent structural insights into the mechanism of ClpP protease regulation by AAA+ chaperones and small molecules. J. Biol. Chem. 2022, 298, 101781.
- Mabanglo, M.F.; Wong, K.S.; Barghash, M.M.; Leung, E.; Chuang, S.H.W.; Ardalan, A.; Majaesic, E.M.; Wong, C.J.; Zhang, S.; Lang, H.; et al. Potent ClpP agonists with anticancer properties bind with improved structural complementarity and alter the mitochondrial N-terminome. Structure 2023, 31, 185–200.e110.
This entry is offline, you can click here to edit this entry!
