Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Oncology
The specificity protein (Sp) transcription factors (TFs) Sp1, Sp2, Sp3 and Sp4 exhibit structural and functional similarities in cancer cells and extensive studies of Sp1 show that it is a negative prognostic factor for patients with multiple tumor types.
- Sp1
- Sp3
- Sp4
- non-oncogene addiction
1. Background
Specificity protein 1 (Sp1) was among the first transcription factors (TFs) identified and is a member of the Sp/Kruppel-like factor (Sp/KLF) family. Members of this family exhibit variable structural domains and functions but all contain conserved zinc fingers in their DNA binding domains that bind GC-rich (Sps) and CACC (KLFs) boxes [1,2,3,4,5,6,7]. Not surprisingly, within the Sp and KLF sub-families there can be some overlap and competition for the same cis-elements, although for many Sp-regulated genes, differences in cell context and levels of expression dictate which Sp transcription factor is active. Among the 9 Sp genes, Sp1-Sp4 are most similar in terms of both structure and function (Figure 1). It should also be pointed out that among Sp1-Sp4, most research has focused on Sp1 and to a lesser extent Sp3 and it is possible that for some genes and pathways, the potential contributions of Sp2 and Sp4 have been understudied. There has been extensive research on the mechanisms of Sp-regulated gene expression, which frequently is observed in genes that lack a TATA box. Many Sp-regulated genes bind and activate gene expression through one or more GC-rich sequences proximal to the start sites where there are ordered assemblies of nuclear cofactors to form a transcriptionally active complex that includes DNA-bound Sp1, Sp3 or Sp4. The composition of transcription complexes includes polymerase II, transcription factor IID (TFIID), TATA box binding protein (TBP) and associated factors (TAFs) and members of the cofactor required for Sp1 activation/mediator (CRISP/MED) complexes [8]. The overall complex is highly variable and both gene- and cell-context-dependent. Moreover, there is also evidence that Sp TFs bind imperfect/variable GC-rich sequences and also interact with distal enhancer sequences, as described for the Topoisomerase IIa promoter [9]. There is a focus on the interactions of Sp TFs with non-coding RNAs and their functions; however, it should also be noted that Sp1 physically interacts with over 55 other proteins [2]. Sp1 function is also influenced by post-transcriptional modifications that include phosphorylation, acetylation, glycosylation and cleavage, and these changes can enhance or inhibit protein stability. Unfortunately, data for Sp3-Sp4 in terms of transcriptional function, post-transcriptional modifications and interactions with other factors have not been extensively investigated.
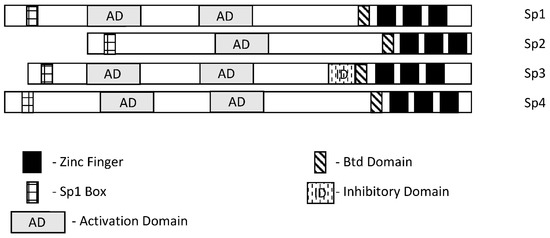
Figure 1. Schematic structures of Sp1, Sp2, Sp3 and Sp4. These transcription factors exhibit several common structural features; however, Sp3 expresses an inhibitory domain that results in gene-specific decreased expression in some cell lines.
2. Sp TFs as Cancer Prognostic Factors
Extensive analysis of tumor and non-tumor tissues has identified many prognostic factors that can be used to predict patient outcomes. Moreover, in some cases, the results dictate the application of specific treatment regimens, and this is particularly true of early-stage breast cancer where expression of estrogen receptor α (ERα, ESR1) in mammary tumors usually results in treatment with endocrine therapies [10]. Table 1 illustrates the important role of Sp1 as a negative prognostic factor for multiple cancers where Sp1 is generally more highly expressed in tumors compared to normal tissue and overexpression is correlated with decreased disease-free patient survival or another negative outcome. With the exception of highly variable results for lung cancer, most tumors overexpress Sp1 (or Sp3) compared to non-tumor tissue and poorer outcomes are observed in patients with tumors overexpressing this TF. In liver cancer, both Sp1 and Sp2 are negative prognostic factors for survival [11,12,13]. In many cases, manuscripts reporting the role of Sp1 as a diagnostic factor are accompanied by laboratory studies showing the pro-oncogenic functional activities of Sp1.
Table 1. Clinical/prognostic Significance of Sp transcription factors.
| Tumor | Sp TF | Prognosis | Refs. |
|---|---|---|---|
| Prostate | Sp1/Sp3/FLIP | Overexpression correlated with a high Gleason score and predicted recurrence | [14] |
| Esophageal squamous cell carcinoma | Sp1 | High Sp1 predicts poor patient survival | [15] |
| Astrocytoma | Sp1 | Poor patient prognosis | [16] |
| Bladder urothelial carcinoma | Sp | Poor clinical outcomes | [17] |
| Glioma | Sp1 | Poor outcomes, higher expression in higher grades, immune invasion | [18,19,20] |
| Head and Neck | Sp3 | Predicted poor survival | [21] |
| Pancreatic | Sp1 (Sp1/LOXL2) | Decreased survival, higher grade, dual prognostic factor (with LOXL2) | [22,23,24] |
| Oral squamous cell carcinoma | Sp1 | Overexpressed and prometastatic | [25] |
| Gastric cancer | Sp1 | Overexpressed, poor prognosis, increased in higher stages | [26,27,28,29,30] |
| Liver cancer | Sp1 | Overexpressed, poor prognosis | [11,12] |
| Colin cancer | Sp1/Sp3 | Overexpressed, decreased survival | [31,32] |
| Breast cancer | Sp1/Par3 | Lower levels/advanced stage, poor prognosis | [33,34,35,36] |
| Lung cancer | Sp1 | Variable prognosis, decreased Sp1 with increasing stage | [37,38,39] |
| Ovarian cancer | Sp1/DANCR | Sp1 overexpression in tumor, correlates with DANCR | [40] |
| Liver cancer | Sp2 | Decreased survival | [13] |
Meta-analysis of multiple studies has also been used to probe the role of Sp1 in gastric cancer, and higher Sp1 expression is correlated with increased depth of invasion and lymph node metastasis, increased TNM staging and Lauren’s classification [41]. A similar meta-analysis approach was used to examine multiple tumor types [42] and similar associations were observed as reported for gastric cancer.
3. Role of Sp in Cell Transformation
Sp1 is clearly a negative prognostic factor for multiple cancers, and this is accompanied by increased expression of Sp transcription factors in tumors compared to non-tumor tissues. These observations suggest that the process that drives the transformation of a normal cell to a tumor cell may also involve Sp transcription factors. This was investigated in a classical study that examined the effects of carcinogen or oncogene-induced transformation of human fibroblasts into fibrosarcoma cells in which the fibrosarcoma, but not the fibroblasts, had the ability to form tumors in athymic nude mice [43,44]. This dramatic change in the phenotype of fibrosarcoma cells compared to the fibroblasts was accompanied by an 8- to 18-fold increased expression of Sp1 protein, which is enhanced during fibroblast cell transformation. Moreover, it was also demonstrated that knockdown of Sp1 in the fibrosarcomas resulted in cells that did not form tumors in athymic nude mice. Other studies show that EGF-induced transformation of bladder epithelial cells and Kras induced transformation of MCFI0A cells also involved Sp1 or an Sp1-regulated gene [45,46]. CYP1B1 also enhanced the proliferation, migration and invasion of MCFI0A and MCF7 cells and this was also accompanied by increased expression of Sp1 and Sp1 regulated genes and silencing or inhibition of Sp1 inhibited CYP1B1-mediated transformation [47].
Arsenic is a carcinogen and considered to be a public health hazard. Exposures of human bronchial epithelial Beas-2B cells to arsenic over a period of several months lead to cell transformation and this was due, in part, to induction DNA methyltransferase 1 (DNMT1) [48]. However, further examination found that arsenic induced Sp1, which in part enhanced DNMT1 expression and loss of miR-199a-5p, which was critical for arsenic-induced transformation. The proposed mechanism involves arsenic-induced Sp1, which in turn activates DNMT1 and suppresses miR-199a-5p. These results demonstrate a role for Sp1 in arsenic-induced transformation of Beas-2B cells; however, the direct effect of Sp1-mediated suppression of miR-199a-5p is unexpected and needs further investigation. Rhabdomyosarcomas (RMS) express high levels of Sp1 compared to non-transformed muscle tissue and RMS cell lines express high levels of Sp1, Sp3 and Sp4. Transformation of human smooth muscles with telomerase, the PAX3-FOXO1 oncogene and NMyc transforms these muscle cell lines; however, expression of only one or two of these factors is not sufficient for transformation [49]. Interestingly, transfection of one or two of these genes dramatically induces expression of Sp1 and Sp3 but not Sp4. This suggests that the process of cell transformation is accompanied by early induction of Sp1 and Sp3 prior to conversion of the muscle cell into a cancer cell [50].
The role of Sp TFs in the process of transformation has also been investigated in cancer stem cells, where they directly regulate genes associated with “stemness” or cooperate with other genes and non-coding RNAs to enhance stemness. At present, there is strong evidence for the role of Sp1 in inducing stemness, and the cooperating factors vary with tumor type. Stemness in breast cancer is maintained by the long non-coding RNA408 (Lnc408)—dependent recruitment of Sp3 to CBY1 gene promoters to inhibit expression of CBY1, which indirectly enhances levels of nuclear β-catenin and β-catenin regulated cancer stem cell-related genes [51]. In gastric cancer, Sp1 regulates expression of leucine-rich repeat-containing receptor 5 (LGR5), a key stem cell factor [52], and in hepatocellular carcinoma, Sp1 induced LncRNA DPPA2 upstream binding RNA (DUBR) [53]. DUBR not only promotes stemness, but also oxaliplatin resistance through an Sp1/DUBR/E2F1-CIP2A axis. The cancer stem-cell-related protein BMI1 is overexpressed in lung cancer and is important for maintaining this phenotype and resistance to pemetrexed [54]. BMI1 also regulates Sp1 expression and knockdown of Sp1 or treatment mithramycin reverses many of the effects of BMI1, including chug resistance. The pro-oncogenic LncRNA HOTAIR interacts with and upregulates Sp1, which induces DNMI1, and transcriptional repression of miR-199a-5p and targeting downregulation of Sp1 or DNMI1 was found to decrease stemness and progression of cutaneous squamous cell carcinoma [55]. In papillary thyroid carcinoma, the LncRNA DOCK9-AS2 interacts with and induces Sp1, which in turn induces β-catenin, which is further induced by DOCK9-AS2 interacting with miR-1972, resulting in increased β-catenin and Wnt signaling [56]. Sp1 is overexpressed in glioblastoma cells [18,19,20] and plays a role in maintaining stemness and drug resistance in this tumor type. It was also reported that ANGPTL4 and Sp4 were overexpressed in GBM and predicted poor patient prognosis [57]. Sp4 also regulates ANGPTL4 and downstream EGFR/AKT/4E-BP1, which is associated with temozolomide resistance and expression of cancer stem cell markers. Drug resistance and stemness in GBM were also associated with Sp1 in another study [58] and in glioma, HDAC/Sp1 regulation of BMI1 enhanced stemness [59]; this exhibited some overlap with lung cancer cells and BMI1 [54].
4. Sp TFs and Regulation of Protein-Encoding Genes in Cancer Cells
In 1983–1984, Tjian and coworkers initially identified Sp1 as a factor that stimulated SV40 early promoter transcription by 40-fold and bound to GC-rich elements in target gene promoters [60,61]. This same group also identified Sp2 as another TF that bound SV40 [60], and approximately a decade later, Sp3 and Sp4 were also characterized [62,63,64,65,66] as a structurally related sub-class of the Sp/KLF family. Subsequent research has demonstrated that Sp1-Sp4 TFs directly regulate or co-regulate thousands of protein-encoding genes associated with cell proliferation, survival, migration and invasion [7]. A detailed study of the role of Sp1, Sp3 and Sp4 in cancer was investigated in multiple cancer cell lines by individual knockdown of the three genes and their combination coupled with analysis of the resulting functional and genomic effects and their overlap [66]. Knockdown of Sp1 (siSp1), Sp3 (siSp3) and Sp4 (siSp4) and their combination (siSp1, 3, 4) decreased growth, increased Annexin V staining (apoptosis) and decreased invasion in A549 lung, MiaPaca2 (pancreatic), SW480 (colon), 786-0 (kidney), SKBR3 (breast), MDA-MB231 (breast), Panc1 (pancreatic) and L3.6 pL (pancreatic) cancer cells. Knockdown efficiencies were high and cell context-dependent differences in functional response potencies were < three-fold for most responses. For most responses, cells deficient in Sp1, Sp3 and Sp4 (triple knockdown) exhibited the highest effect on growth inhibition, induction of Annexin V staining and inhibition of invasion; however, the magnitude of the differences between single and triple knockdown was relatively modest. These results indicate that Sp1, Sp3 and Sp4 individually regulate proliferation, survival and invasion of cancer cells and the loss of one of these TFs is not compensated or rescued by the other two. One possible explanation is that Sp1, Sp3 and Sp4 cooperatively regulate many of the same pro-oncogenic genes and loss of a single TF compromises any possible rescue by the other two.
The highly invasive Panc1 pancreatic cancer cell line was used as a model to investigate the differential expression of genes after knockdown of Sp1, Sp3 and Sp4. Figure 2 illustrates the number of DEGs after knockdown of Sp1, Sp3 and Sp4, including 3532, 4826 and 4232 genes, respectively. Further analysis shows that the common DEGs after knockdown of Sp1/Sp3, Sp1/Sp4 and Sp3/Sp4 were 1113, 1140 and 2753, respectively, indicating that pairs of the three Sp TFs regulated a relatively high percentage of genes in common. This was particularly true for Sp3/Sp4, in which 2753 genes were commonly regulated by both transcription factors, which includes 57 and 64% of all Sp3 and Sp4 regulated genes, respectively. This would suggest that particularly for Sp3 and Sp4 and also the other pairs (Sp1/Sp3, Sp1/Sp4), there may be significant cooperative regulation of genes that requires more than one Sp TF. As demonstrated in Figure 2 and Figure 3, Sp1, Sp3 and Sp4 regulate expression of several thousand genes, with many of them associated with cancer proliferation, survival, and migration/invasion. Moreover, the three transcription factors also regulate genes in common and also genes that are Sp- specific and vary with cell context. Sp (Sp1, Sp3 and Sp4) regulated genes include epidermal growth factor receptor 1 (EGFR), other tyrosine kinases, cMyc, bcl2, survivin, vascular endothelial growth factor receptors (VEGFR1 and VEGFR2), matrix metalloproteinases and many other genes.
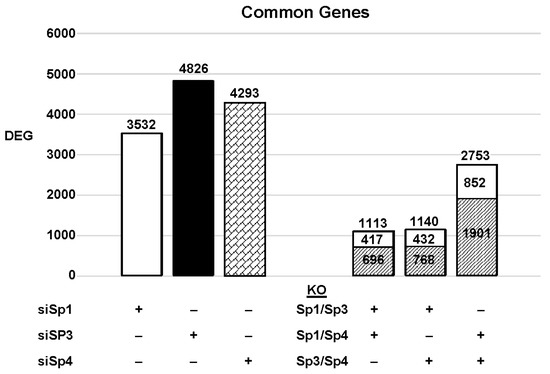
Figure 2. Sp knockdown and changes in gene expression. Panc1 cells were transfected with siRNAs, and after Sp knockdown, the changes in gene expression and the genes commonly induced/repressed by siSp1/siSp3, siSp1/siSp4, and siSp4/siSp3 were determined ( : decreased and
: decreased and  : increased expression in the double knockout groups).
: increased expression in the double knockout groups).
 : decreased and
: decreased and  : increased expression in the double knockout groups).
: increased expression in the double knockout groups).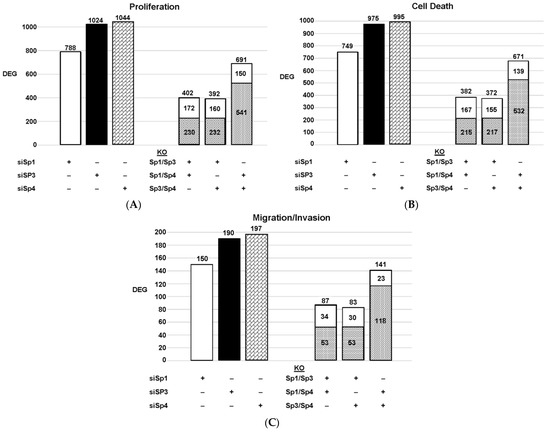
Figure 3. Effects of Sp knockdown by RNAi: IPA analysis of the differentially expressed genes in Panc1 cells associated with cell proliferation (A), Annexin V staining (B), and invasion (C). In these same samples, the common genes observed after knockdown of Sp1/Sp3, Sp1/Sp4 and Sp3/Sp4 are given. ( : decreased and
: decreased and  : increased expression in the double knockout group).
: increased expression in the double knockout group).
 : decreased and
: decreased and  : increased expression in the double knockout group).
: increased expression in the double knockout group).Since Sp TF regulate genes associated with cell proliferation, survival, and invasion, we used ingenuity pathway analysis (IPA) to analyze DEGs for each pathway after knockdown of individual Sps and their combination. The relative expressions of DEGs were determined and the results are illustrated in Figure 3. The patterns of DEGs associated with Panc1 cell proliferation, survival, and invasion after knockdown of Sp1, Sp3 and Sp4 were similar; however, the number of genes involved followed the order of proliferation ≥cell death > invasion. In addition, the pattern of the number of DEGs commonly expressed by Sp1/Sp3, Sp1/Sp4 and Sp3/Sp4 associated with cell proliferation, survival and invasion was higher than that observed for the total genes. The percentage of common genes/total genes was the highest for Sp3/Sp4, where the percentages were 67%, 68% and 74% (Sp3), and 66%, 67% and 72% (Sp4) for cell proliferation, survival, and invasion respectively. Casual IPA analysis also confirmed by their z scores (>2.0 or <−2.0) that the DEGs in each group were strongly associated with the functional responses.
There is evidence from the large number of publications that not only do Sp1, Sp3 and Sp4 regulate pro-oncogenic pathways and genes, but there are also reports that Sp2 performs similar functions [13,67,68]. For example, Sp2 knockdown in hepatocellular carcinoma cells decreases cell migration, proliferation and survival of hepatocellular carcinoma cells and this is due, in part, to decreasing the expression of the TRIB3 gene [13]. Additionally, Sp2-dependent suppression carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) [67] and overexpression of Sp2 increase susceptibility to wound- and carcinogen-induced tumorigenesis [68]. Thus, Sp1-Sp4 regulation of protein-encoding genes plays an important role in cell transformation and tumorigenesis.
This entry is adapted from the peer-reviewed paper 10.3390/ijms24065164
This entry is offline, you can click here to edit this entry!
