The application of artificial intelligence (AI) is accelerating the paradigm shift towards patient-tailored brain tumor management, achieving optimal onco-functional balance for each individual. AI-based models can positively impact different stages of the diagnostic and therapeutic process. Although the histological investigation will remain difficult to replace, in the near future the radiomic approach will allow a complementary, repeatable and non-invasive characterization of the lesion, assisting oncologists and neurosurgeons in selecting the best therapeutic option and the correct molecular target in chemotherapy. AI-driven tools are already playing an important role in surgical planning, delimiting the extent of the lesion (segmentation) and its relationships with the brain structures, thus allowing precision brain surgery as radical as reasonably acceptable to preserve the quality of life. Finally, AI-assisted models allow the prediction of complications, recurrences and therapeutic response, suggesting the most appropriate follow-up.
- artificial intelligence
- brain tumors
- glioblastoma
- deep learning
- prognosis prediction
1. Introduction
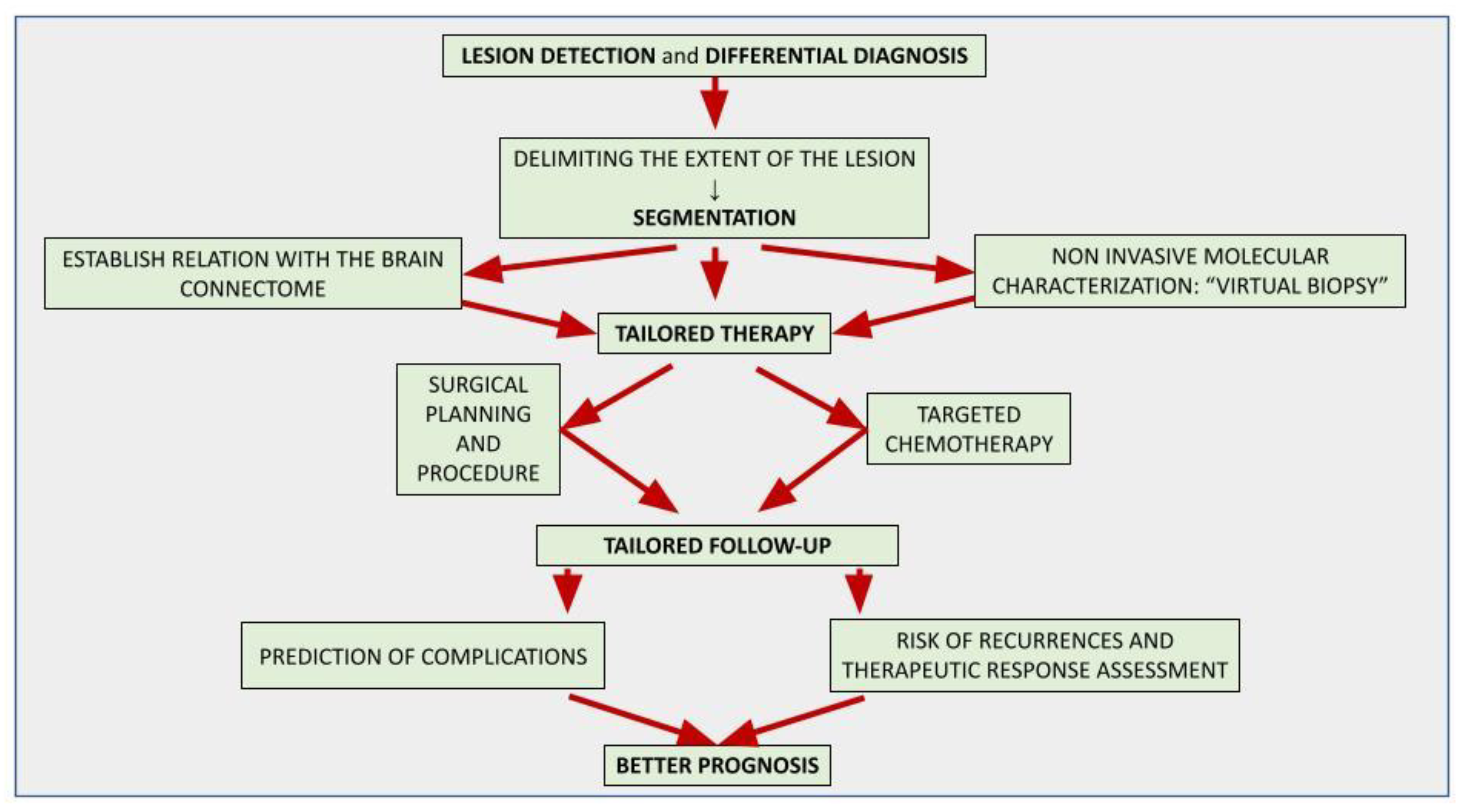
2. An Introduction to Artificial Intelligence and Related Concepts
2.1. Artificial Intelligence (AI)
2.2. Radiomics
3. Lesion Detection and Differential Diagnosis
4. Tumor Characterization
This entry is adapted from the peer-reviewed paper 10.3390/curroncol30030203
References
- Abdel Razek, A.A.K.; Alksas, A.; Shehata, M.; AbdelKhalek, A.; Abdel Baky, K.; El-Baz, A.; Helmy, E. Clinical Applications of Artificial Intelligence and Radiomics in Neuro-Oncology Imaging. Insights Imaging 2021, 12, 152.
- Wesseling, P.; Capper, D. WHO 2016 Classification of Gliomas. Neuropathol. Appl. Neurobiol. 2018, 44, 139–150.
- Jiang, H.; Cui, Y.; Wang, J.; Lin, S. Impact of Epidemiological Characteristics of Supratentorial Gliomas in Adults Brought about by the 2016 World Health Organization Classification of Tumors of the Central Nervous System. Oncotarget 2017, 8, 20354–20361.
- Ceravolo, I.; Barchetti, G.; Biraschi, F.; Gerace, C.; Pampana, E.; Pingi, A.; Stasolla, A. Early Stage Glioblastoma: Retrospective Multicentric Analysis of Clinical and Radiological Features. Radiol. Med. 2021, 126, 1468–1476.
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 WHO Classification of Tumours of the Central Nervous System. Acta Neuropathol. 2007, 114, 97–109.
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathol. 2016, 131, 803–820.
- Capper, D.; Jones, D.T.W.; Sill, M.; Hovestadt, V.; Schrimpf, D.; Sturm, D.; Koelsche, C.; Sahm, F.; Chavez, L.; Reuss, D.E.; et al. DNA Methylation-Based Classification of Central Nervous System Tumours. Nature 2018, 555, 469–474.
- Luger, A.-L.; König, S.; Samp, P.F.; Urban, H.; Divé, I.; Burger, M.C.; Voss, M.; Franz, K.; Fokas, E.; Filipski, K.; et al. Molecular Matched Targeted Therapies for Primary Brain Tumors—A Single Center Retrospective Analysis. J. Neurooncol. 2022, 159, 243–259.
- Di Bonaventura, R.; Montano, N.; Giordano, M.; Gessi, M.; Gaudino, S.; Izzo, A.; Mattogno, P.P.; Stumpo, V.; Caccavella, V.M.; Giordano, C.; et al. Reassessing the Role of Brain Tumor Biopsy in the Era of Advanced Surgical, Molecular, and Imaging Techniques—A Single-Center Experience with Long-Term Follow-Up. J. Pers. Med. 2021, 11, 909.
- Singh, G.; Manjila, S.; Sakla, N.; True, A.; Wardeh, A.H.; Beig, N.; Vaysberg, A.; Matthews, J.; Prasanna, P.; Spektor, V. Radiomics and Radiogenomics in Gliomas: A Contemporary Update. Br. J. Cancer 2021, 125, 641–657.
- Vagvala, S.; Guenette, J.P.; Jaimes, C.; Huang, R.Y. Imaging Diagnosis and Treatment Selection for Brain Tumors in the Era of Molecular Therapeutics. Cancer Imaging 2022, 22, 19.
- Rudie, J.D.; Rauschecker, A.M.; Bryan, R.N.; Davatzikos, C.; Mohan, S. Emerging Applications of Artificial Intelligence in Neuro-Oncology. Radiology 2019, 290, 607–618.
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting More Information from Medical Images Using Advanced Feature Analysis. Eur. J. Cancer 2012, 48, 441–446.
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J.W.L. Artificial Intelligence in Radiology. Nat. Rev. Cancer 2018, 18, 500–510.
- Scapicchio, C.; Gabelloni, M.; Barucci, A.; Cioni, D.; Saba, L.; Neri, E. A Deep Look into Radiomics. Radiol. Med. 2021, 126, 1296–1311.
- Mayerhoefer, M.E.; Materka, A.; Langs, G.; Häggström, I.; Szczypiński, P.; Gibbs, P.; Cook, G. Introduction to Radiomics. J. Nucl. Med. 2020, 61, 488–495.
- Irmici, G.; Cè, M.; Caloro, E.; Khenkina, N.; Della Pepa, G.; Ascenti, V.; Martinenghi, C.; Papa, S.; Oliva, G.; Cellina, M. Chest X-Ray in Emergency Radiology: What Artificial Intelligence Applications Are Available? Diagnostics 2023, 13, 216.
- Soda, P.; D’Amico, N.C.; Tessadori, J.; Valbusa, G.; Guarrasi, V.; Bortolotto, C.; Akbar, M.U.; Sicilia, R.; Cordelli, E.; Fazzini, D.; et al. AIforCOVID: Predicting the Clinical Outcomes in Patients with COVID-19 Applying AI to Chest-X-Rays. An Italian Multicentre Study. Med. Image Anal. 2021, 74, 102216.
- Pope, W.B. Brain Metastases: Neuroimaging. Handb. Clin. Neurol. 2018, 149, 89–112.
- Cellina, M.; Cè, M.; Irmici, G.; Ascenti, V.; Khenkina, N.; Toto-Brocchi, M.; Martinenghi, C.; Papa, S.; Carrafiello, G. Artificial Intelligence in Lung Cancer Imaging: Unfolding the Future. Diagnostics 2022, 12, 2644.
- Park, J.E. Artificial Intelligence in Neuro-Oncologic Imaging: A Brief Review for Clinical Use Cases and Future Perspectives. Brain Tumor Res. Treat. 2022, 10, 69.
- Park, Y.W.; Jun, Y.; Lee, Y.; Han, K.; An, C.; Ahn, S.S.; Hwang, D.; Lee, S.-K. Robust Performance of Deep Learning for Automatic Detection and Segmentation of Brain Metastases Using Three-Dimensional Black-Blood and Three-Dimensional Gradient Echo Imaging. Eur. Radiol. 2021, 31, 6686–6695.
- Voicu, I.P.; Pravatà, E.; Panara, V.; Navarra, R.; Mattei, P.A.; Caulo, M. Differentiating Solitary Brain Metastases from High-Grade Gliomas with MR: Comparing Qualitative versus Quantitative Diagnostic Strategies. Radiol. Med. 2022, 127, 891–898.
- Bauer, A.H.; Erly, W.; Moser, F.G.; Maya, M.; Nael, K. Differentiation of Solitary Brain Metastasis from Glioblastoma Multiforme: A Predictive Multiparametric Approach Using Combined MR Diffusion and Perfusion. Neuroradiology 2015, 57, 697–703.
- Romano, A.; Moltoni, G.; Guarnera, A.; Pasquini, L.; Di Napoli, A.; Napolitano, A.; Espagnet, M.C.R.; Bozzao, A. Single Brain Metastasis versus Glioblastoma Multiforme: A VOI-Based Multiparametric Analysis for Differential Diagnosis. Radiol. Med. 2022, 127, 490–497.
- Swinburne, N.C.; Schefflein, J.; Sakai, Y.; Oermann, E.K.; Titano, J.J.; Chen, I.; Tadayon, S.; Aggarwal, A.; Doshi, A.; Nael, K. Machine Learning for Semi-automated Classification of Glioblastoma, Brain Metastasis and Central Nervous System Lymphoma Using Magnetic Resonance Advanced Imaging. Ann. Transl. Med. 2019, 7, 232.
- Upadhyay, N.; Waldman, A.D. Conventional MRI Evaluation of Gliomas. Br. J. Radiol. 2011, 84, S107–S111.
- Skogen, K.; Schulz, A.; Helseth, E.; Ganeshan, B.; Dormagen, J.B.; Server, A. Texture Analysis on Diffusion Tensor Imaging: Discriminating Glioblastoma from Single Brain Metastasis. Acta Radiol. 2019, 60, 356–366.
- Han, Y.; Zhang, L.; Niu, S.; Chen, S.; Yang, B.; Chen, H.; Zheng, F.; Zang, Y.; Zhang, H.; Xin, Y.; et al. Differentiation between Glioblastoma Multiforme and Metastasis from the Lungs and Other Sites Using Combined Clinical/Routine MRI Radiomics. Front. Cell Dev. Biol. 2021, 9, 710461.
- Nayak, L.; Lee, E.Q.; Wen, P.Y. Epidemiology of Brain Metastases. Curr. Oncol. Rep. 2012, 14, 48–54.
- Ortiz-Ramón, R.; Larroza, A.; Ruiz-España, S.; Arana, E.; Moratal, D. Classifying Brain Metastases by Their Primary Site of Origin Using a Radiomics Approach Based on Texture Analysis: A Feasibility Study. Eur. Radiol. 2018, 28, 4514–4523.
- Barajas, R.F.; Politi, L.S.; Anzalone, N.; Schöder, H.; Fox, C.P.; Boxerman, J.L.; Kaufmann, T.J.; Quarles, C.C.; Ellingson, B.M.; Auer, D.; et al. Consensus Recommendations for MRI and PET Imaging of Primary Central Nervous System Lymphoma: Guideline Statement from the International Primary CNS Lymphoma Collaborative Group (IPCG). Neuro Oncol. 2021, 23, 1056–1071.
- Tang, Y.Z.; Booth, T.C.; Bhogal, P.; Malhotra, A.; Wilhelm, T. Imaging of Primary Central Nervous System Lymphoma. Clin. Radiol. 2011, 66, 768–777.
- Cai, Q.; Fang, Y.; Young, K.H. Primary Central Nervous System Lymphoma: Molecular Pathogenesis and Advances in Treatment. Transl. Oncol. 2019, 12, 523–538.
- Stadlbauer, A.; Marhold, F.; Oberndorfer, S.; Heinz, G.; Buchfelder, M.; Kinfe, T.M.; Meyer-Bäse, A. Radiophysiomics: Brain Tumors Classification by Machine Learning and Physiological MRI Data. Cancers 2022, 14, 2363.
- Ucuzal, H.; Yasar, S.; Colak, C. Classification of Brain Tumor Types by Deep Learning with Convolutional Neural Network on Magnetic Resonance Images Using a Developed Web-Based Interface. In Proceedings of the 2019 3rd International Symposium on Multidisciplinary Studies and Innovative Technologies (ISMSIT), Ankara, Turkey, 11–13 October 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1–5.
- Adu, K.; Yu, Y.; Cai, J.; Tashi, N. Dilated Capsule Network for Brain Tumor Type Classification via MRI Segmented Tumor Region. In Proceedings of the 2019 IEEE International Conference on Robotics and Biomimetics (ROBIO), Dali, China, 6–8 December 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 942–947.
- Afshar, P.; Plataniotis, K.N.; Mohammadi, A. Capsule Networks for Brain Tumor Classification Based on MRI Images and Coarse Tumor Boundaries. In Proceedings of the ICASSP 2019–2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1368–1372.
- Sunderland, G.J.; Jenkinson, M.D.; Zakaria, R. Surgical Management of Posterior Fossa Metastases. J. Neurooncol. 2016, 130, 535–542.
- She, D.; Yang, X.; Xing, Z.; Cao, D. Differentiating Hemangioblastomas from Brain Metastases Using Diffusion-Weighted Imaging and Dynamic Susceptibility Contrast-Enhanced Perfusion-Weighted MR Imaging. Am. J. Neuroradiol. 2016, 37, 1844–1850.
- Payabvash, S.; Aboian, M.; Tihan, T.; Cha, S. Machine Learning Decision Tree Models for Differentiation of Posterior Fossa Tumors Using Diffusion Histogram Analysis and Structural MRI Findings. Front. Oncol. 2020, 10, 71.
- Lin, X.; Yu, W.-Y.; Liauw, L.; Chander, R.J.; Soon, W.E.; Lee, H.Y.; Tan, K. Clinicoradiologic Features Distinguish Tumefactive Multiple Sclerosis from CNS Neoplasms. Neurol. Clin. Pract. 2017, 7, 53–64.
- Verma, R.K.; Wiest, R.; Locher, C.; Heldner, M.R.; Schucht, P.; Raabe, A.; Gralla, J.; Kamm, C.P.; Slotboom, J.; Kellner-Weldon, F. Differentiating Enhancing Multiple Sclerosis Lesions, Glioblastoma, and Lymphoma with Dynamic Texture Parameters Analysis: A Feasibility Study. Med. Phys. 2017, 44, 4000–4008.
- Han, Y.; Yang, Y.; Shi, Z.; Zhang, A.; Yan, L.; Hu, Y.; Feng, L.; Ma, J.; Wang, W.; Cui, G. Distinguishing Brain Inflammation from Grade II Glioma in Population without Contrast Enhancement: A Radiomics Analysis Based on Conventional MRI. Eur. J. Radiol. 2021, 134, 109467.
- Qian, Z.; Li, Y.; Wang, Y.; Li, L.; Li, R.; Wang, K.; Li, S.; Tang, K.; Zhang, C.; Fan, X.; et al. Differentiation of Glioblastoma from Solitary Brain Metastases Using Radiomic Machine-Learning Classifiers. Cancer Lett. 2019, 451, 128–135.
- Bae, S.; An, C.; Ahn, S.S.; Kim, H.; Han, K.; Kim, S.W.; Park, J.E.; Kim, H.S.; Lee, S.-K. Robust Performance of Deep Learning for Distinguishing Glioblastoma from Single Brain Metastasis Using Radiomic Features: Model Development and Validation. Sci. Rep. 2020, 10, 12110.
- Wiestler, B.; Kluge, A.; Lukas, M.; Gempt, J.; Ringel, F.; Schlegel, J.; Meyer, B.; Zimmer, C.; Förster, S.; Pyka, T.; et al. Multiparametric MRI-Based Differentiation of WHO Grade II/III Glioma and WHO Grade IV Glioblastoma. Sci. Rep. 2016, 6, 35142.
- Zhang, X.; Yan, L.-F.; Hu, Y.-C.; Li, G.; Yang, Y.; Han, Y.; Sun, Y.-Z.; Liu, Z.-C.; Tian, Q.; Han, Z.-Y.; et al. Optimizing a Machine Learning Based Glioma Grading System Using Multi-Parametric MRI Histogram and Texture Features. Oncotarget 2017, 8, 47816–47830.
- Kim, M.; Jung, S.Y.; Park, J.E.; Jo, Y.; Park, S.Y.; Nam, S.J.; Kim, J.H.; Kim, H.S. Diffusion- and Perfusion-Weighted MRI Radiomics Model May Predict Isocitrate Dehydrogenase (IDH) Mutation and Tumor Aggressiveness in Diffuse Lower Grade Glioma. Eur. Radiol. 2020, 30, 2142–2151.
- Akkus, Z.; Ali, I.; Sedlář, J.; Agrawal, J.P.; Parney, I.F.; Giannini, C.; Erickson, B.J. Predicting Deletion of Chromosomal Arms 1p/19q in Low-Grade Gliomas from MR Images Using Machine Intelligence. J. Digit. Imaging 2017, 30, 469–476.
- Cho, H.; Lee, S.; Kim, J.; Park, H. Classification of the Glioma Grading Using Radiomics Analysis. PeerJ 2018, 6, e5982.
- Tian, Q.; Yan, L.-F.; Zhang, X.; Zhang, X.; Hu, Y.-C.; Han, Y.; Liu, Z.-C.; Nan, H.-Y.; Sun, Q.; Sun, Y.-Z.; et al. Radiomics Strategy for Glioma Grading Using Texture Features from Multiparametric MRI. J. Magn. Reson. Imaging 2018, 48, 1518–1528.
- Mzoughi, H.; Njeh, I.; Wali, A.; Slima, M.B.; BenHamida, A.; Mhiri, C.; Mahfoudhe, K.B. Deep Multi-Scale 3D Convolutional Neural Network (CNN) for MRI Gliomas Brain Tumor Classification. J. Digit. Imaging 2020, 33, 903–915.
- Chang, P.; Grinband, J.; Weinberg, B.D.; Bardis, M.; Khy, M.; Cadena, G.; Su, M.-Y.; Cha, S.; Filippi, C.G.; Bota, D.; et al. Deep-Learning Convolutional Neural Networks Accurately Classify Genetic Mutations in Gliomas. Am. J. Neuroradiol. 2018, 39, 1201–1207.
- Meng, L.; Zhang, R.; Fa, L.; Zhang, L.; Wang, L.; Shao, G. ATRX Status in Patients with Gliomas: Radiomics Analysis. Medicine 2022, 101, e30189.
- Ren, Y.; Zhang, X.; Rui, W.; Pang, H.; Qiu, T.; Wang, J.; Xie, Q.; Jin, T.; Zhang, H.; Chen, H.; et al. Noninvasive Prediction of IDH1 Mutation and ATRX Expression Loss in Low-Grade Gliomas Using Multiparametric MR Radiomic Features. J. Magn. Reson. Imaging 2019, 49, 808–817.
- Alentorn, A.; Duran-Peña, A.; Pingle, S.C.; Piccioni, D.E.; Idbaih, A.; Kesari, S. Molecular profiling of gliomas: Potential therapeutic implications. Expert Rev. Anticancer Ther. 2015, 15, 955–962.
- Haubold, J.; Hosch, R.; Parmar, V.; Glas, M.; Guberina, N.; Catalano, O.A.; Pierscianek, D.; Wrede, K.; Deuschl, C.; Forsting, M.; et al. Fully Automated MR Based Virtual Biopsy of Cerebral Gliomas. Cancers 2021, 13, 6186.
- Shboul, Z.A.; Chen, J.; Iftekharuddin, K.M. Prediction of Molecular Mutations in Diffuse Low-Grade Gliomas Using MR Imaging Features. Sci. Rep. 2020, 10, 3711.
- Calabrese, E.; Rudie, J.D.; Rauschecker, A.M.; Villanueva-Meyer, J.E.; Clarke, J.L.; Solomon, D.A.; Cha, S. Combining Radiomics and Deep Convolutional Neural Network Features from Preoperative MRI for Predicting Clinically Relevant Genetic Biomarkers in Glioblastoma. Neuro-Oncol. Adv. 2022, 4, vdac060.
