Autophagy–the lysosomal degradation of cytoplasm–plays a central role in cellular homeostasis and protects cells from potentially harmful agents that may accumulate in the cytoplasm, including pathogens, protein aggregates, and dysfunctional organelles. This process is initiated by the formation of a phagophore membrane, which wraps around a portion of cytoplasm or cargo and closes to form a double-membrane autophagosome. Upon the fusion of the autophagosome with a lysosome, the sequestered material is degraded by lysosomal hydrolases in the resulting autolysosome. Several alternative membrane sources of autophagosomes have been proposed, including the plasma membrane, endosomes, mitochondria, endoplasmic reticulum, lipid droplets, hybrid organelles, and de novo synthesis.
- autophagy
- autophagosome
- endosome
1. Introduction
2. The Preautophagosomal Structure as Autophagosome Generator in Budding Yeast
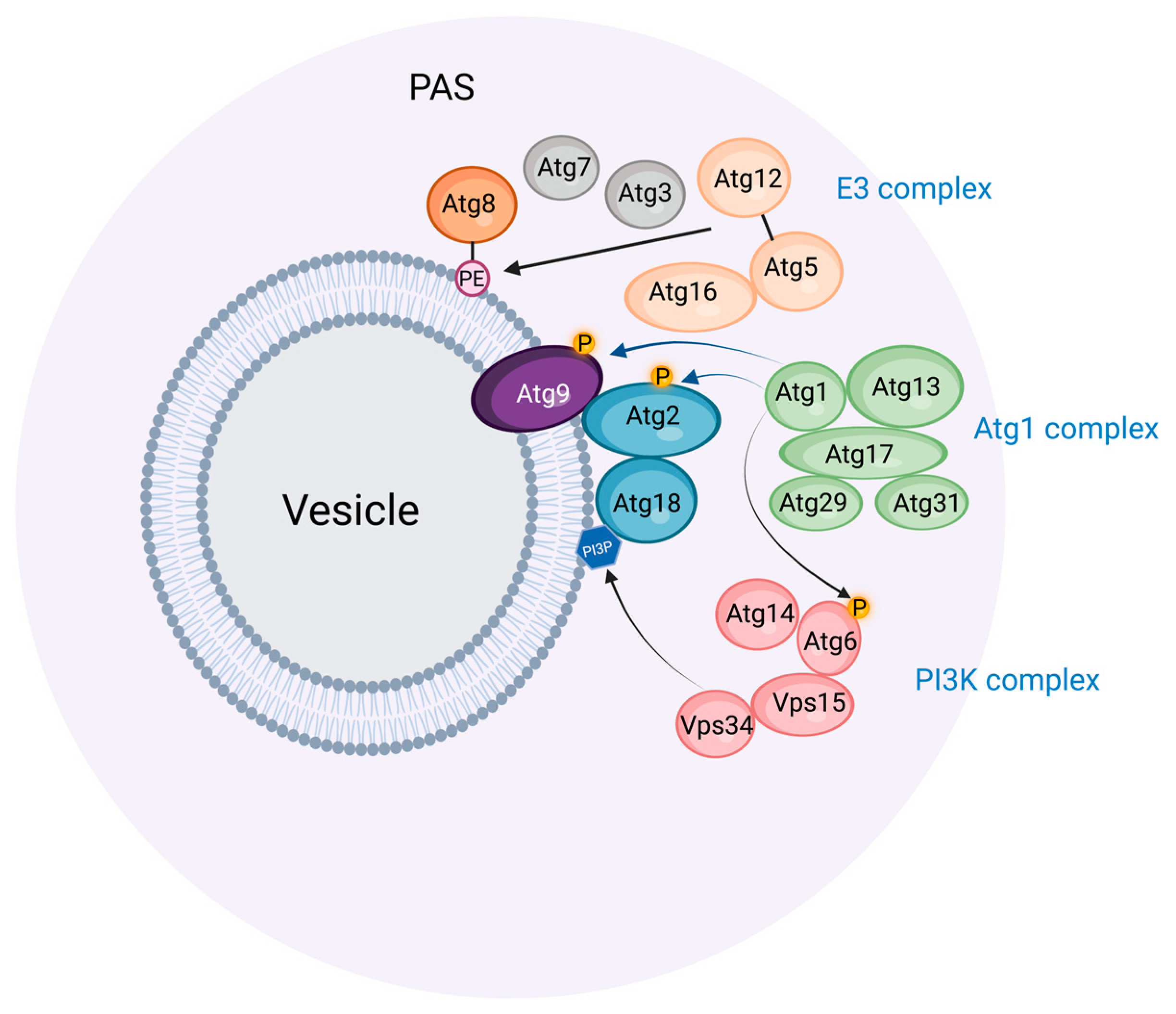
3. Similarities and Differences between Budding Yeast and Mammals in Autophagosome Biogenesis
4. The Endoplasmic Reticulum as Source of Phagophores
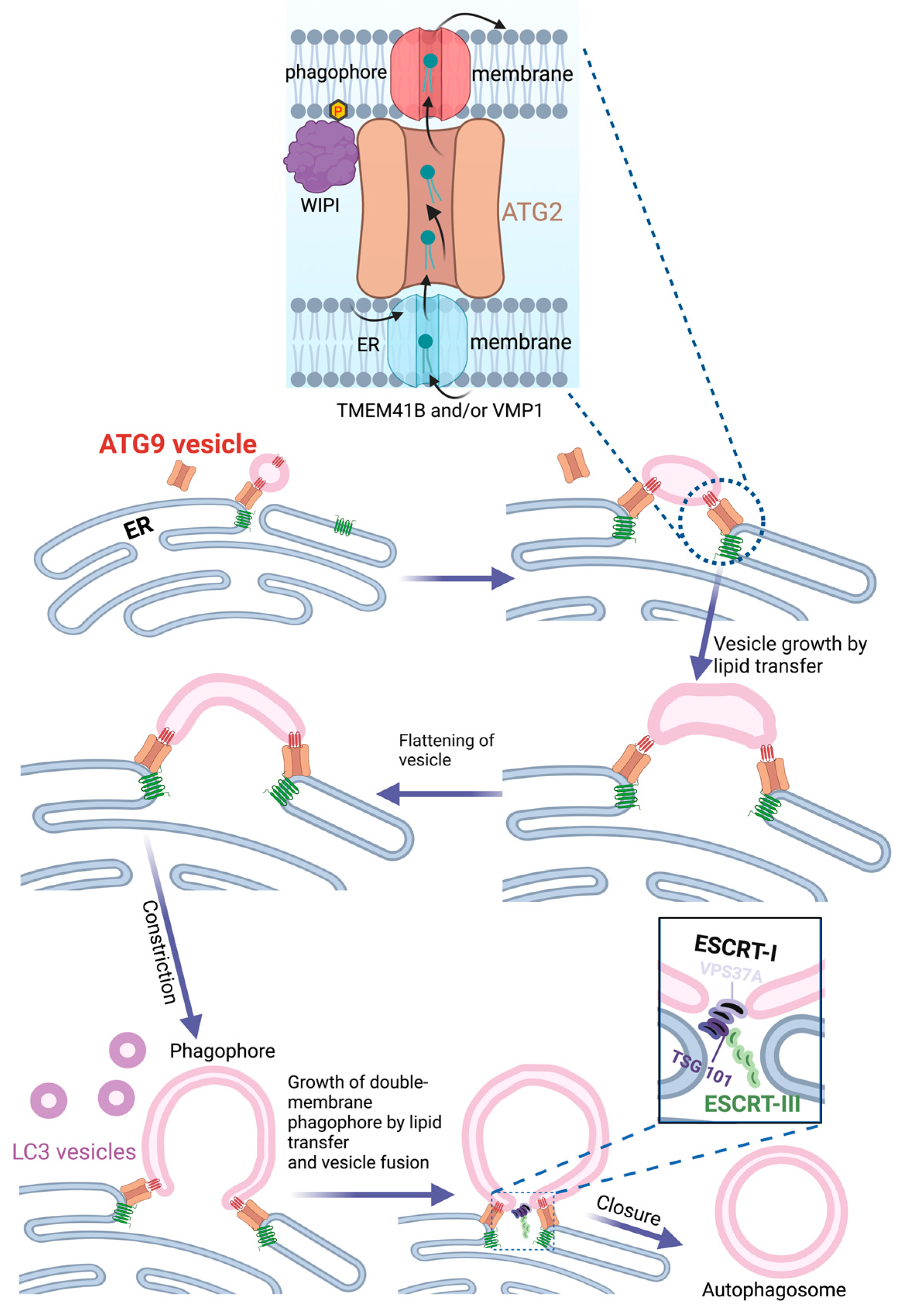
5. COPII Vesicles and ER Exit Sites in Phagophore Biogenesis
6. Mitochondria as Source of Phagophore Membranes
7. Contact Sites between ER and Mitochondria as Source of Phagophores
This entry is adapted from the peer-reviewed paper 10.3390/cells12040668
References
- Nakatogawa, H.; Suzuki, K.; Kamada, Y.; Ohsumi, Y. Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat. Rev. Mol. Cell Biol. 2009, 10, 458–467.
- Dikic, I.; Elazar, Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 349–364.
- Seglen, P.O.; Gordon, P.B.; Holen, I. Non-selective autophagy. Semin. Cell Biol. 1990, 1, 441–448.
- Takeshige, K.; Baba, M.; Tsuboi, S.; Noda, T.; Ohsumi, Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 1992, 119, 301–311.
- Gudmundsson, S.R.; Kallio, K.A.; Vihinen, H.; Jokitalo, E.; Ktistakis, N.; Eskelinen, E.-L. Morphology of Phagophore Precursors by Correlative Light-Electron Microscopy. Cells 2022, 11, 3080.
- Berg, T.O.; Fengsrud, M.; Strømhaug, P.E.; Berg, T.; Seglen, P.O. Isolation and characterization of rat liver amphisomes—Evidence for fusion of autophagosomes with both early and late endosomes. J. Biol. Chem. 1998, 273, 21883–21892.
- Yu, L.; McPhee, C.K.; Zheng, L.; Mardones, G.A.; Rong, Y.; Peng, J.; Mi, N.; Zhao, Y.; Liu, Z.; Wan, F.; et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 2010, 465, 942–946.
- Melia, T.J.; Lystad, A.H.; Simonsen, A. Autophagosome biogenesis: From membrane growth to closure. J. Cell Biol. 2020, 219, e202002085.
- Suzuki, K.; Ohsumi, Y. Current knowledge of the pre-autophagosomal structure (PAS). FEBS Lett. 2010, 584, 1280–1286.
- Hollenstein, D.; Gómez-Sánchez, R.; Ciftci, A.M.A.; Kriegenburg, F.; Mari, M.; Torggler, R.; Licheva, M.; Reggiori, F.; Kraft, C. Vac8 spatially confines autophagosome formation at the vacuole. J. Cell Sci. 2019, 132, jcs235002.
- Fujioka, Y.; Alam, J.M.; Noshiro, D.; Mouri, K.; Ando, T.; Okada, Y.; May, A.I.; Knorr, R.L.; Suzuki, K.; Ohsumi, Y.; et al. Phase separation organizes the site of autophagosome formation. Nature 2020, 578, 301–305.
- Yamamoto, H.; Fujioka, Y.; Suzuki, S.W.; Noshiro, D.; Suzuki, H.; Kondo-Kakuta, C.; Kimura, Y.; Hirano, H.; Ando, T.; Noda, N.N.; et al. The Intrinsically Disordered Protein Atg13 Mediates Supramolecular Assembly of Autophagy Initiation Complexes. Dev. Cell 2016, 38, 86–99.
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The Role of Atg Proteins in Autophagosome Formation. Annu. Rev. Cell Dev. Biol. 2011, 27, 107–132.
- Kihara, A.; Noda, T.; Ishihara, N.; Ohsumi, Y. Two Distinct Vps34 Phosphatidylinositol 3–Kinase Complexes Function in Autophagy and Carboxypeptidase Y Sorting inSaccharomyces cerevisiae. J. Cell Biol. 2001, 152, 519–530.
- Kotani, T.; Kirisako, H.; Koizumi, M.; Ohsumi, Y.; Nakatogawa, H. The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc. Natl. Acad. Sci. USA 2018, 115, 10363–10368.
- Obara, K.; Ohsumi, Y. Dynamics and function of PtdIns(3)P in autophagy. Autophagy 2008, 4, 952–954.
- Ramirez, S.C.; Gómez-Sánchez, R.; Verlhac, P.; Hardenberg, R.; Margheritis, E.; Cosentino, K.; Reggiori, F.; Ungermann, C. Atg9 interactions via its transmembrane domains are required for phagophore expansion during autophagy. Autophagy 2022, 1–20.
- Cebollero, E.; Van Der Vaart, A.; Reggiori, F. Understanding phosphatidylinositol-3-phosphate dynamics during autophagosome biogenesis. Autophagy 2012, 8, 1868–1870.
- Noda, N.N.; Inagaki, F. Mechanisms of Autophagy. Annu. Rev. Biophys. 2015, 44, 101–122.
- Nakatogawa, H.; Ichimura, Y.; Ohsumi, Y. Atg8, a Ubiquitin-like Protein Required for Autophagosome Formation, Mediates Membrane Tethering and Hemifusion. Cell 2007, 130, 165–178.
- Maruyama, T.; Alam, J.M.; Fukuda, T.; Kageyama, S.; Kirisako, H.; Ishii, Y.; Shimada, I.; Ohsumi, Y.; Komatsu, M.; Kanki, T.; et al. Membrane perturbation by lipidated Atg8 underlies autophagosome biogenesis. Nat. Struct. Mol. Biol. 2021, 28, 583–593.
- Hayashi-Nishino, M.; Fujita, N.; Noda, T.; Yamaguchi, A.; Yoshimori, T.; Yamamoto, A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 2009, 11, 1433–1437.
- Ylä-Anttila, P.; Vihinen, H.; Jokitalo, E.; Eskelinen, E.-L. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 2009, 5, 1180–1185.
- Takahashi, S.; Saito, C.; Koyama-Honda, I.; Mizushima, N. Quantitative 3D correlative light and electron microscopy of organelle association during autophagy. Cell Struct. Funct. 2022, 47, 89–99.
- Axe, E.L.; Walker, S.A.; Manifava, M.; Chandra, P.; Roderick, H.L.; Habermann, A.; Griffiths, G.; Ktistakis, N.T. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008, 182, 685–701.
- Kishi-Itakura, C.; Koyama-Honda, I.; Itakura, E.; Mizushima, N. Ultrastructural analysis of autophagosome organization using mammalian autophagy-deficient cells. J. Cell Sci. 2014, 127, 4089–4102.
- Matoba, K.; Kotani, T.; Tsutsumi, A.; Tsuji, T.; Mori, T.; Noshiro, D.; Sugita, Y.; Nomura, N.; Iwata, S.; Ohsumi, Y.; et al. Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat. Struct. Mol. Biol. 2020, 27, 1185–1193.
- Maeda, S.; Yamamoto, H.; Kinch, L.N.; Garza, C.M.; Takahashi, S.; Otomo, C.; Grishin, N.V.; Forli, S.; Mizushima, N.; Otomo, T. Structure, lipid scrambling activity and role in autophagosome formation of ATG9A. Nat. Struct. Mol. Biol. 2020, 27, 1194–1201.
- Valverde, D.P.; Yu, S.; Boggavarapu, V.; Kumar, N.; Lees, J.A.; Walz, T.; Reinisch, K.M.; Melia, T.J. ATG2 transports lipids to promote autophagosome biogenesis. J. Cell Biol. 2019, 218, 1787–1798.
- Ghanbarpour, A.; Valverde, D.P.; Melia, T.J.; Reinisch, K.M. A model for a partnership of lipid transfer proteins and scramblases in membrane expansion and organelle biogenesis. Proc. Natl. Acad. Sci. USA 2021, 118, e2101562118.
- van Vliet, A.R.; Chiduza, G.N.; Maslen, S.L.; Pye, V.E.; Joshi, D.; De Tito, S.; Jefferies, H.B.; Christodoulou, E.; Roustan, C.; Punch, E.; et al. ATG9A and ATG2A form a heteromeric complex essential for autophagosome formation. Mol. Cell 2022, 82, 4324–4339.e8.
- Melia, T.J.; Reinisch, K.M. A possible role for VPS13-family proteins in bulk lipid transfer, membrane expansion and organelle biogenesis. J. Cell Sci. 2022, 135, jcs259357.
- Muñoz-Braceras, S.; Calvo, R.; Escalante, R. TipC and the chorea-acanthocytosis protein VPS13A regulate autophagy in Dictyostelium and human HeLa cells. Autophagy 2015, 11, 918–927.
- Kumar, N.; Leonzino, M.; Hancock-Cerutti, W.; Horenkamp, F.A.; Li, P.; Lees, J.A.; Wheeler, H.; Reinisch, K.M.; De Camilli, P. VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 2018, 217, 3625–3639.
- Chen, S.; Mari, M.; Parashar, S.; Liu, D.; Cui, Y.; Reggiori, F.; Novick, P.J.; Ferro-Novick, S. Vps13 is required for the packaging of the ER into autophagosomes during ER-phagy. Proc. Natl. Acad. Sci. USA 2020, 117, 18530–18539.
- Fengsrud, M.; Erichsen, E.S.; Berg, T.O.; Raiborg, C.; Seglen, P.O. Ultrastructural characterization of the delimiting membranes of isolated autophagosomes and amphisomes by freeze-fracture electron microscopy. Eur. J. Cell Biol. 2000, 79, 871–882.
- Ravussin, A.; Brech, A.; Tooze, S.A.; Stenmark, H. The phosphatidylinositol 3-phosphate-binding protein SNX4 controls ATG9A recycling and autophagy. J. Cell Sci. 2021, 134, jcs250670.
- Antonny, B.; Schekman, R. ER export: Public transportation by the COPII coach. Curr. Opin. Cell Biol. 2001, 13, 438–443.
- Ishihara, N.; Hamasaki, M.; Yokota, S.; Suzuki, K.; Kamada, Y.; Kihara, A.; Yoshimori, T.; Noda, T.; Ohsumi, Y. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell 2001, 12, 3690–3702.
- Graef, M.; Friedman, J.; Graham, C.; Babu, M.; Nunnari, J. ER exit sites are physical and functional core autophagosome biogenesis components. Mol. Biol. Cell 2013, 24, 2918–2931.
- Tan, D.; Cai, Y.; Wang, J.; Zhang, J.; Menon, S.; Chou, H.-T.; Ferro-Novick, S.; Reinisch, K.M.; Walz, T. The EM structure of the TRAPPIII complex leads to the identification of a requirement for COPII vesicles on the macroautophagy pathway. Proc. Natl. Acad. Sci. USA 2013, 110, 19432–19437.
- Scrima, S.; Tiberti, M.; Campo, A.; Corcelle-Termeau, E.; Judith, D.; Foged, M.M.; Clemmensen, K.K.B.; Tooze, S.A.; Jäättelä, M.; Maeda, K.; et al. Unraveling membrane properties at the organelle-level with LipidDyn. Comput. Struct. Biotechnol. J. 2022, 20, 3604–3614.
- Imai, K.; Hao, F.; Fujita, N.; Tsuji, Y.; Oe, Y.; Araki, Y.; Hamasaki, M.; Noda, T.; Yoshimori, T. Atg9A trafficking through the recycling endosomes is required for autophagosome formation. J. Cell Sci. 2016, 129, 3781–3791.
- Tremel, S.; Ohashi, Y.; Morado, D.R.; Bertram, J.; Perisic, O.; Brandt, L.T.L.; von Wrisberg, M.-K.; Chen, Z.A.; Maslen, S.L.; Kovtun, O.; et al. Structural basis for VPS34 kinase activation by Rab1 and Rab5 on membranes. Nat. Commun. 2021, 12, 1564.
- Wang, J.; Menon, S.; Yamasaki, A.; Chou, H.-T.; Walz, T.; Jiang, Y.; Ferro-Novick, S. Ypt1 recruits the Atg1 kinase to the preautophagosomal structure. Proc. Natl. Acad. Sci. USA 2013, 110, 9800–9805.
- Hailey, D.W.; Rambold, A.S.; Satpute-Krishnan, P.; Mitra, K.; Sougrat, R.; Kim, P.K.; Lippincott-Schwartz, J. Mitochondria Supply Membranes for Autophagosome Biogenesis during Starvation. Cell 2010, 141, 656–667.
- Pickles, S.; Vigié, P.; Youle, R.J. Mitophagy and Quality Control Mechanisms in Mitochondrial Maintenance. Curr. Biol. 2018, 28, R170–R185.
- Vance, J.E. Thematic Review Series: Glycerolipids. Phosphatidylserine and phosphatidylethanolamine in mammalian cells: Two metabolically related aminophospholipids. J. Lipid Res. 2008, 49, 1377–1387.
- Hamasaki, M.; Furuta, N.; Matsuda, A.; Nezu, A.; Yamamoto, A.; Fujita, N.; Oomori, H.; Noda, T.; Haraguchi, T.; Hiraoka, Y.; et al. Autophagosomes form at ER–mitochondria contact sites. Nature 2013, 495, 389–393.
- De Brito, O.M.; Scorrano, L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 2008, 456, 605–610.
- Karanasios, E.; Walker, S.A.; Okkenhaug, H.; Manifava, M.; Hummel, E.; Zimmermann, H.; Ahmed, Q.; Domart, M.-C.; Collinson, L.; Ktistakis, N.T. Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nat. Commun. 2016, 7, 12420.
