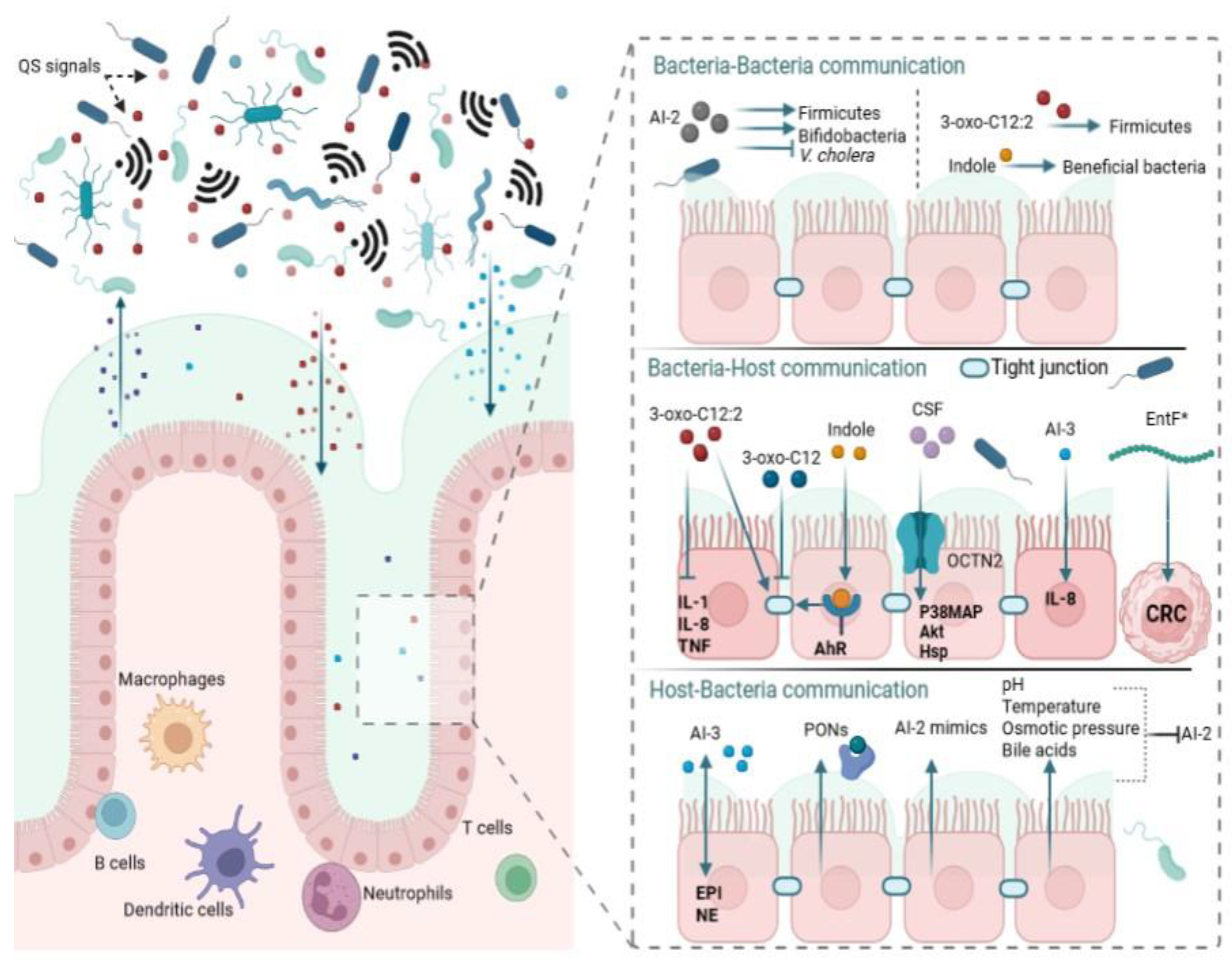
An imbalance in gut microbiota, termed dysbiosis, has been shown to affect host health. Several factors, including dietary changes, have been reported to cause dysbiosis with its associated pathologies that include inflammatory bowel disease, cancer, obesity, depression, and autism. Quorum sensing (QS) is a complex network of cell–cell communication that is mediated by small diffusible molecules known as autoinducers (AIs). Using AIs, bacteria interact with one another and coordinate their gene expression based on their population density for the benefit of the whole community or one group over another. Bacteria that cannot synthesize their own AIs secretly “listen” to the signals produced by other bacteria, a phenomenon known as “eavesdropping”. AIs impact gut microbiota equilibrium by mediating intra- and interspecies interactions as well as interkingdom communication.
- gut
- bacteria
- quorum sensing
- autoinducers
- normobiosis
- dysbiosis
1. Introduction
2. Normobiosis
2.1. Mode of Delivery
2.2. Diet
2.3. Genetics
2.4. Geographical Location
2.5. Other Factors
3. Quorum Sensing
4. Different Quorum Sensing Signals in the Gut
4.1. Autoinducing Peptides
4.2. N-Acyl Homoserine Lactones
4.3. Autoinducer-2
4.4. Autoinducer-3
5. Role of QS in Normobiosis
5.1. Intraspecies QS and Normobiosis
5.2. Interspecies QS and Normobiosis
5.3. Interkingdom QS and Normobiosis
6. Conclusions
This entry is adapted from the peer-reviewed paper 10.3390/ijms24043722
References
- Landman, C.; Quévrain, E. Le microbiote intestinal: Description, rôle et implication physiopathologique. La Rev. Médecine Interne 2016, 37, 418–423.
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836.
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms 2019, 7, 14.
- Doré, J.; Corthier, G. The human intestinal microbiota. Gastroentérol. Clin. Biol. 2010, 34, S7–S15.
- Lepage, P. Le microbiome digestif humain: Interactions avec l’hôte et dysfonctions. Rev. Mal. Respir. 2017, 34, 1085–1090.
- Lane, E.R.; Zisman, T.L.; Suskind, D.L. The microbiota in inflammatory bowel disease: Current and therapeutic insights. J. Inflamm. Res. 2017, 10, 63–73.
- Ronan, V.; Yeasin, R.; Claud, E.C. Childhood Development and the Microbiome—The Intestinal Microbiota in Maintenance of Health and Development of Disease During Childhood Development. Gastroenterology 2021, 160, 495–506.
- Martin, R.; Makino, H.; Cetinyurek Yavuz, A.; Ben-Amor, K.; Roelofs, M.; Ishikawa, E.; Kubota, H.; Swinkels, S.; Sakai, T.; Oishi, K.; et al. Early-Life Events, Including Mode of Delivery and Type of Feeding, Siblings and Gender, Shape the Developing Gut Microbiota. PLoS ONE 2016, 11, e0158498.
- Houghteling, P.D.; Walker, W.A. Why Is Initial Bacterial Colonization of the Intestine Important to Infants’ and Children’s Health? J. Pediatr. Gastroenterol. Nutr. 2015, 60, 294–307.
- Ley, R.E.; Peterson, D.A.; Gordon, J.I. Ecological and Evolutionary Forces Shaping Microbial Diversity in the Human Intestine. Cell 2006, 124, 837–848.
- Marques, T.M.; Wall, R.; Ross, R.P.; Fitzgerald, G.F.; Ryan, C.A.; Stanton, C. Programming infant gut microbiota: Influence of dietary and environmental factors. Curr. Opin. Biotechnol. 2010, 21, 149–156.
- Arrieta, M.-C.; Stiemsma, L.T.; Amenyogbe, N.; Brown, E.M.; Finlay, B. The Intestinal Microbiome in Early Life: Health and Disease. Front. Immunol. 2014, 5, 427.
- Satokari, R.; Grönroos, T.; Laitinen, K.; Salminen, S.; Isolauri, E. Bifidobacterium and Lactobacillus DNA in the human placenta. Lett. Appl. Microbiol. 2009, 48, 8–12.
- Jiménez, E.; Fernández, L.; Marín, M.L.; Martín, R.; Odriozola, J.M.; Nueno-Palop, C.; Narbad, A.; Olivares, M.; Xaus, J.; Rodríguez, J.M. Isolation of Commensal Bacteria from Umbilical Cord Blood of Healthy Neonates Born by Cesarean Section. Curr. Microbiol. 2005, 51, 270–274.
- Jiménez, E.; Marín, M.L.; Martín, R.; Odriozola, J.M.; Olivares, M.; Xaus, J.; Fernández, L.; Rodríguez, J.M. Is meconium from healthy newborns actually sterile? Res. Microbiol. 2008, 159, 187–193.
- Gaufin, T.; Tobin, N.H.; Aldrovandi, G.M. The importance of the microbiome in pediatrics and pediatric infectious diseases. Curr. Opin. Pediatr. 2018, 30, 117–124.
- Makino, H.; Kushiro, A.; Ishikawa, E.; Muylaert, D.; Kubota, H.; Sakai, T.; Oishi, K.; Martin, R.; Ben Amor, K.; Oozeer, R.; et al. Transmission of Intestinal Bifidobacterium longum subsp. longum Strains from Mother to Infant, Determined by Multilocus Sequencing Typing and Amplified Fragment Length Polymorphism. Appl. Environ. Microbiol. 2011, 77, 6788–6793.
- Makino, H.; Kushiro, A.; Ishikawa, E.; Kubota, H.; Gawad, A.; Sakai, T.; Oishi, K.; Martin, R.; Ben-Amor, K.; Knol, J.; et al. Mother-to-Infant Transmission of Intestinal Bifidobacterial Strains Has an Impact on the Early Development of Vaginally Delivered Infant’s Microbiota. PLoS ONE 2013, 8, e78331.
- Milani, C.; Mancabelli, L.; Lugli, G.A.; Duranti, S.; Turroni, F.; Ferrario, C.; Mangifesta, M.; Viappiani, A.; Ferretti, P.; Gorfer, V.; et al. Exploring Vertical Transmission of Bifidobacteria from Mother to Child. Appl. Environ. Microbiol. 2015, 81, 7078–7087.
- Biasucci, G.; Benenati, B.; Morelli, L.; Bessi, E.; Boehm, G. Cesarean Delivery May Affect the Early Biodiversity of Intestinal Bacteria. J. Nutr. 2008, 138, 1796S–1800S.
- Shao, Y.; Forster, S.C.; Tsaliki, E.; Vervier, K.; Strang, A.; Simpson, N.; Kumar, N.; Stares, M.D.; Rodger, A.; Brocklehurst, P.; et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 2019, 574, 117–121.
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 2015, 17, 852.
- Lewis, Z.T.; Mills, D.A. Differential Establishment of Bifidobacteria in the Breastfed Infant Gut. In Nestle Nutrition Institute Workshop Series; Karger Publishers: Basel, Switzerland, 2017; Volume 88, pp. 149–160.
- Wampach, L.; Heintz-Buschart, A.; Hogan, A.; Muller, E.E.L.; Narayanasamy, S.; Laczny, C.C.; Hugerth, L.W.; Bindl, L.; Bottu, J.; Andersson, A.F.; et al. Colonization and Succession within the Human Gut Microbiome by Archaea, Bacteria, and Microeukaryotes during the First Year of Life. Front. Microbiol. 2017, 8, 738.
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227.
- Koenig, J.E.; Spor, A.; Scalfone, N.; Fricker, A.D.; Stombaugh, J.; Knight, R.; Angenent, L.T.; Ley, R.E. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. USA 2011, 108, 4578–4585.
- Palmer, C.; Bik, E.M.; DiGiulio, D.B.; Relman, D.A.; Brown, P.O. Development of the Human Infant Intestinal Microbiota. PLoS Biol. 2007, 5, e177.
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T.; et al. Human Genetics Shape the Gut Microbiome. Cell 2014, 159, 789–799.
- Prescott, C.A.; Kendler, K.S. Twin Study Design. Alcohol Health Res. World 1995, 19, 200–205.
- Goodrich, J.K.; Davenport, E.R.; Waters, J.L.; Clark, A.G.; Ley, R.E. Cross-species comparisons of host genetic associations with the microbiome. Science 2016, 352, 532–535.
- Goodrich, J.K.; Davenport, E.R.; Beaumont, M.; Jackson, M.A.; Knight, R.; Ober, C.; Spector, T.D.; Bell, J.T.; Clark, A.G.; Ley, R.E. Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host Microbe 2016, 19, 731–743.
- Xie, H.; Guo, R.; Zhong, H.; Feng, Q.; Lan, Z.; Qin, B.; Ward, K.J.; Jackson, M.A.; Xia, Y.; Chen, X.; et al. Shotgun Metagenomics of 250 Adult Twins Reveals Genetic and Environmental Impacts on the Gut Microbiome. Cell Syst. 2016, 3, 572–584.
- Abeles, S.R.; Jones, M.B.; Santiago-Rodriguez, T.M.; Ly, M.; Klitgord, N.; Yooseph, S.; Nelson, K.E.; Pride, D.T. Microbial diversity in individuals and their household contacts following typical antibiotic courses. Microbiome 2016, 4, 39.
- Gupta, V.; Kumar, R.; Sood, U.; Singhvi, N. Reconciling Hygiene and Cleanliness: A New Perspective from Human Microbiome. Indian J. Microbiol. 2020, 60, 37–44.
- Freestone, P.P.E.; Sandrini, S.M.; Haigh, R.D.; Lyte, M. Microbial endocrinology: How stress influences susceptibility to infection. Trends Microbiol. 2008, 16, 55–64.
- Moreira, C.G.; Russell, R.; Mishra, A.A.; Narayanan, S.; Ritchie, J.M.; Waldor, M.K.; Curtis, M.M.; Winter, S.E.; Weinshenker, D.; Sperandio, V. Bacterial Adrenergic Sensors Regulate Virulence of Enteric Pathogens in the Gut. MBio 2016, 7, 00826-16.
- Badal, V.D.; Vaccariello, E.D.; Murray, E.R.; Yu, K.E.; Knight, R.; Jeste, D.V.; Nguyen, T.T. The Gut Microbiome, Aging, and Longevity: A Systematic Review. Nutrients 2020, 12, 3759.
- O’Toole, G.A. Classic spotlight: Quorum sensing and the multicellular life of unicellular organisms. J. Bacteriol. 2016, 198, 601.
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427.
- Nealson, K.H.; Platt, T.; Hastings, J.W. Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 1970, 104, 313–322.
- Fuqua, W.C.; Winans, S.C.; Greenberg, E.P. Quorum sensing in bacteria: The LuxR-LuxI family of cell density- responsive transcriptional regulators. J. Bacteriol. 1994, 176, 269–275.
- Bassler, B.L.; Greenberg, E.P.; Stevens, A.M. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 1997, 179, 4043–4045.
- Chen, X.; Schauder, S.; Potier, N.; Van Dorsselaer, A.; Pelczer, I.; Bassler, B.L.; Hughson, F.M. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 2002, 415, 545–549.
- Xavier, K.B.; Bassler, B.L. Interference with AI-2-mediated bacterial cell-cell communication. Nature 2005, 437, 750–753.
- An, J.H.; Goo, E.; Kim, H.; Seo, Y.S.; Hwang, I. Bacterial quorum sensing and metabolic slowing in a cooperative population. Proc. Natl. Acad. Sci. USA 2014, 111, 14912–14917.
- Mukherjee, S.; Bassler, B.L. Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 2019, 17, 371–382.
- Azimi, S.; Klementiev, A.D.; Whiteley, M.; Diggle, S.P. Bacterial Quorum Sensing during Infection. Annu. Rev. Microbiol. 2020, 74, 201–219.
- Wang, S.; Payne, G.F.; Bentley, W.E. Quorum Sensing Communication: Molecularly Connecting Cells, Their Neighbors, and even Devices. Annu. Rev. Chem. Biomol. Eng. 2020, 11, 447–468.
- Smith, R.S.; Iglewski, B.H.P. aeruginosa quorum-sensing systems and virulence. Curr. Opin. Microbiol. 2003, 6, 56–60.
- Galloway, W.R.J.D.; Hodgkinson, J.T.; Bowden, S.D.; Welch, M.; Spring, D.R. Quorum sensing in Gram-negative bacteria: Small-molecule modulation of AHL and AI-2 quorum sensing pathways. Chem. Rev. 2011, 111, 28–67.
- Prescott, R.D.; Decho, A.W. Flexibility and Adaptability of Quorum Sensing in Nature. Trends Microbiol. 2020, 28, 436–444.
- Chen, L.; Wen, Y.M. The role of bacterial biofilm in persistent infections and control strategies. Int. J. Oral Sci. 2011, 3, 66–73.
- Harmsen, M.; Yang, L.; Pamp, S.J.; Tolker-Nielsen, T. An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol. Med. Microbiol. 2010, 59, 253–268.
- Kim, C.S.; Gatsios, A.; Cuesta, S.; Lam, Y.C.; Wei, Z.; Chen, H.; Russell, R.M.; Shine, E.E.; Wang, R.; Wyche, T.P.; et al. Characterization of Autoinducer-3 Structure and Biosynthesis in E. coli. ACS Cent. Sci. 2020, 6, 197–206.
- Whitehead, N.A.; Barnard, A.M.L.; Slater, H.; Simpson, N.J.L.; Salmond, G.P.C. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 2001, 25, 365–404.
- Rai, N.; Rai, R.; Venkatesh, K.V. Quorum Sensing in Competence and Sporulation. In Quorum Sensing vs Quorum Quenching: A Battle with No End in Sight; Kalia, V.C., Ed.; Springer: New Delhi, India, 2015; pp. 61–64. ISBN 978-81-322-1982-8.
- Perchat, S.; Talagas, A.; Poncet, S.; Lazar, N.; Li de la Sierra-Gallay, I.; Gohar, M.; Lereclus, D.; Nessler, S. How Quorum Sensing Connects Sporulation to Necrotrophism in Bacillus thuringiensis. PLoS Pathog. 2016, 12, 1005779.
- Winson, M.K.; Swift, S.; Fish, L.; Throup, J.P.; Jørgensen, F.; Chhabra, S.R.; Bycroft, B.W.; Williams, P.; Stewart, G.S.A.B. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol. Lett. 1998, 163, 185–192.
- Goo, E.; An, J.H.; Kang, Y.; Hwang, I. Control of bacterial metabolism by quorum sensing. Trends Microbiol. 2015, 23, 567–576.
- Zhu, L.; Chen, T.; Xu, L.; Zhou, Z.; Feng, W.; Liu, Y.; Chen, H. Effect and mechanism of quorum sensing on horizontal transfer of multidrug plasmid RP4 in BAC biofilm. Sci. Total Environ. 2020, 698, 134236.
- El-Sayed, A.K.; Hothersall, J.; Thomas, C.M. Quorum-sensing-dependent regulation of biosynthesis of the polyketide antiobiotic mupirocin in Pseudomonas fluorescens NCIMB 10586. Microbiology 2001, 147, 2127–2139.
- Ha, J.H.; Hauk, P.; Cho, K.; Eo, Y.; Ma, X.; Stephens, K.; Cha, S.; Jeong, M.; Suh, J.Y.; Sintim, H.O.; et al. Evidence of link between quorum sensing and sugar metabolism in Escherichia coli revealed via cocrystal structures of LsrK and HPr. Sci. Adv. 2018, 4, aar7063.
- Pena, R.T.; Blasco, L.; Ambroa, A.; González-Pedrajo, B.; Fernández-García, L.; López, M.; Bleriot, I.; Bou, G.; García-Contreras, R.; Wood, T.K.; et al. Relationship between quorum sensing and secretion systems. Front. Microbiol. 2019, 10, 1100.
- Kang, Y.; Goo, E.; Kim, J.; Hwang, I. Critical role of quorum sensing-dependent glutamate metabolism in homeostatic osmolality and outer membrane vesiculation in Burkholderia glumae. Sci. Rep. 2017, 7, 44195.
- Barnard, A.M.L.; Bowden, S.D.; Burr, T.; Coulthurst, S.J.; Monson, R.E.; Salmond, G.P.C. Quorum sensing, virulence and secondary metabolite production in plant soft-rotting bacteria. Philos. Trans. R. Soc. B Biol. Sci. 2007, 362, 1165–1183.
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199.
- Moura-Alves, P.; Puyskens, A.; Stinn, A.; Klemm, M.; Guhlich-Bornhof, U.; Dorhoi, A.; Furkert, J.; Kreuchwig, A.; Protze, J.; Lozza, L.; et al. Host monitoring of quorum sensing during Pseudomonas aeruginosa infection. Science 2019, 366, aaw1629.
- Landman, C.; Grill, J.-P.; Mallet, J.-M.; Marteau, P.; Humbert, L.; Le Balc’h, E.; Maubert, M.-A.; Perez, K.; Chaara, W.; Brot, L.; et al. Inter-kingdom effect on epithelial cells of the N-Acyl homoserine lactone 3-oxo-C12:2, a major quorum-sensing molecule from gut microbiota. PLoS ONE 2018, 13, e0202587.
- Li, Q.; Ren, Y.; Fu, X. Inter-kingdom signaling between gut microbiota and their host. Cell. Mol. Life Sci. 2019, 76, 2383–2389.
- Ismail, A.S.; Valastyan, J.S.; Bassler, B.L. A Host-Produced Autoinducer-2 Mimic Activates Bacterial Quorum Sensing. Cell Host Microbe 2016, 19, 470–480.
- Coquant, G.; Aguanno, D.; Pham, S.; Grellier, N.; Thenet, S.; Carrière, V.; Grill, J.-P.; Seksik, P. Gossip in the gut: Quorum sensing, a new player in the host-microbiota interactions. World J. Gastroenterol. 2021, 27, 7247–7270.
- Verbeke, F.; De Craemer, S.; Debunne, N.; Janssens, Y.; Wynendaele, E.; Van de Wiele, C.; De Spiegeleer, B. Peptides as quorum sensing molecules: Measurement techniques and obtained levels in vitro and in vivo. Front. Neurosci. 2017, 11, 183.
- Ziemichód, A.; Skotarczak, B. QS-systems communication of Gram-positive bacterial cells. Acta Biol. 2017, 24, 51–56.
- Thoendel, M.; Kavanaugh, J.S.; Flack, C.E.; Horswill, A.R. Peptide signaling in the Staphylococci. Chem. Rev. 2011, 111, 117–151.
- Wei, Y.; Ng, W.L.; Cong, J.; Bassler, B.L. Ligand and antagonist driven regulation of the Vibrio cholerae quorum-sensing receptor CqsS. Mol. Microbiol. 2012, 83, 1095–1108.
- Liu, Z.; Zhang, L.; Wang, J.; Li, Y.; Chang, Y.; Huang, X.; Duan, J.; Ai, Y.; Zeng, X.; Guo, J. Virtual Screening and Biological Evaluation of Anti-Biofilm Agents Targeting LuxS in the Quorum Sensing System. Nat. Prod. Commun. 2021, 16, 1934578X2110196.
- Schuster, M.; Joseph Sexton, D.; Diggle, S.P.; Peter Greenberg, E. Acyl-homoserine lactone quorum sensing: From evolution to application. Annu. Rev. Microbiol. 2013, 67, 43–63.
- Papenfort, K.; Bassler, B.L. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588.
- Parsek, M.R.; Val, D.L.; Hanzelka, B.L.; Cronan, J.E.; Greenberg, E.P. Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. USA 1999, 96, 4360–4365.
- Bassler, B.L.; Wright, M.; Showalter, R.E.; Silverman, M.R. Intercellular signalling in Vibrio harveyi: Sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 1993, 9, 773–786.
- Bassler, B.L.; Wright, M.; Silverman, M.R. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: Sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 1994, 13, 273–286.
- Schauder, S.; Bassler, B.L. The languages of bacteria. Genes Dev. 2001, 15, 1468–1480.
- Surette, M.G.; Miller, M.B.; Bassler, B.L. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: A new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 1999, 96, 1639–1644.
- Schauder, S.; Shokat, K.; Surette, M.G.; Bassler, B.L. The LuxS family of bacterial autoinducers: Biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 2001, 41, 463–476.
- Cornell, K.A.; Swarts, W.E.; Barry, R.D.; Riscoe, M.K. Characterization of Recombinant Eschericha coli Analysis of Enzymatic Activity and Substrate Specificity drugs which specifically inhibit microbial growth while leaving the human host unaffected. adenosine (MTA) and S-adenosylhomocysteine (SAH), bypr. Biochem. Biophys. Res. Commun. 1996, 732, 724–732.
- Wnuk, S.F.; Lalama, J.; Robert, J.; Garmendia, C.A. Novel S-ribosylhomocysteine analogues as potential inhibitors of LuxS enzyme. Nucl. Nucl. Nucleic Acids 2007, 26, 1051–1055.
- Pei, D.; Zhu, J. Mechanism of action of S-ribosylhomocysteinase (LuxS). Curr. Opin. Chem. Biol. 2004, 8, 492–497.
- Sperandio, V.; Torres, A.G.; Jarvis, B.; Nataro, J.P.; Kaper, J.B. Bacteria-host communication: The language of hormones. Proc. Natl. Acad. Sci. USA 2003, 100, 8951–8956.
- Walters, M.; Sperandio, V. Autoinducer 3 and epinephrine signaling in the kinetics of locus of enterocyte effacement gene expression in enterohemorrhagic Escherichia coli. Infect. Immun. 2006, 74, 5445–5455.
- Kendall, M.M.; Rasko, D.A.; Sperandio, V. Global effects of the cell-to-cell signaling molecules autoinducer-2, autoinducer-3, and epinephrine in a luxS mutant of enterohemorrhagic Escherichia coli. Infect. Immun. 2007, 75, 4875–4884.
- Hernandez, D.E.; Sintim, H.O. Quorum Sensing Autoinducer-3 Finally Yields to Structural Elucidation. ACS Cent. Sci. 2020, 6, 93–96.
- Clarke, M.B.; Hughes, D.T.; Zhu, C.; Boedeker, E.C.; Sperandio, V. The QseC sensor kinase: A bacterial adrenergic receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 10420–10425.
- Halang, P.; Toulouse, C.; Geißel, B.; Michel, B.; Flauger, B.; Müller, M.; Voegele, R.T.; Stefanski, V.; Steuber, J. Response of Vibrio cholerae to the Catecholamine Hormones Epinephrine and Norepinephrine. J. Bacteriol. 2015, 197, 3769–3778.
- Swearingen, M.C.; Sabag-Daigle, A.; Ahmer, B.M.M. Are there acyl-homoserine lactones within mammalian intestines? J. Bacteriol. 2013, 195, 173–179.
- Landman, C.; Besse, A.; Maubert, M.; Brot, L.; Humbert, L.; Cosnes, J.; Beaugerie, L.; Trugnan, G.; Sokol, H.; Rainteau, D.; et al. Sa1804 Quorum Sensing Driven by N-Acyl-Homoserine Lactone in Inflammatory Bowel Diseases Associated Dysbiosis. Gastroenterology 2013, 144, S-310.
- Le Balc’h, E.; Landman, C.; Tauziet, E.; Brot, L.; Quevrain, E.; Rainteau, D.; Grill, J.-P.; Thenet, S.; Seksik, P. P780 3-oxo-C12:2-HSL, a new N-acyl-homoserine lactone identified in gut ecosystem exerts an anti-inflammatory effect and does not modify paracellular permeability. J. Crohn’s Colitis 2017, 11, S479–S480.
- Aguanno, D.; Coquant, G.; Peyrottes, A.; Landman, C.; Brot, L.; Mallet, J.-M.; Quevrain, E.; Carriere, V.; Osinski, C.; Leturque, A.; et al. Mo1113—3-Oxo-C12:2 N-Acyl Homoserine Lactone, a Quorum Sensing Molecule from the Gut Microbiota: A New Player to Control Gut Inflammation. Gastroenterology 2019, 156, S-710.
- Aguanno, D.; Coquant, G.; Postal, B.G.; Osinski, C.; Wieckowski, M.; Stockholm, D.; Grill, J.P.; Carrière, V.; Seksik, P.; Thenet, S. The intestinal quorum sensing 3-oxo-C12:2 Acyl homoserine lactone limits cytokine-induced tight junction disruption. Tissue Barriers 2020, 8, 183287.
- Thompson, J.A.; Oliveira, R.A.; Djukovic, A.; Ubeda, C.; Xavier, K.B. Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 2015, 10, 1861–1871.
- Wang, G.; Wilson, T.J.M.; Jiang, Q.; Taylor, D.E. Spontaneous Mutations That Confer Antibiotic Resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 2001, 45, 727–733.
- Hsiao, A.; Ahmed, A.M.S.; Subramanian, S.; Griffin, N.W.; Drewry, L.L.; Petri, W.A.; Haque, R.; Ahmed, T.; Gordon, J.I. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature 2014, 515, 423–426.
- Lee, J.-H.; Lee, J. Indole as an intercellular signal in microbial communities. FEMS Microbiol. Rev. 2010, 34, 426–444.
- Lee, J.; Zhang, X.-S.; Hegde, M.; Bentley, W.E.; Jayaraman, A.; Wood, T.K. Indole cell signaling occurs primarily at low temperatures in Escherichia coli. ISME J. 2008, 2, 1007–1023.
- Yang, J.; Chawla, R.; Rhee, K.Y.; Gupta, R.; Manson, M.D.; Jayaraman, A.; Lele, P.P. Biphasic chemotaxis of Escherichia coli to the microbiota metabolite indole. Proc. Natl. Acad. Sci. USA 2020, 117, 6114–6120.
- Kumar, A.; Sperandio, V. Indole Signaling at the Host-Microbiota-Pathogen Interface. MBio 2019, 10, 19.
- Bansal, T.; Englert, D.; Lee, J.; Hegde, M.; Wood, T.K.; Jayaraman, A. Differential effects of epinephrine, norepinephrine, and indole on Escherichia coli O157:H7 chemotaxis, colonization, and gene expression. Infect. Immun. 2007, 75, 4597–4607.
- Lee, J.; Bansal, T.; Jayaraman, A.; Bentley, W.E.; Wood, T.K. Enterohemorrhagic Escherichia coli biofilms are inhibited by 7-hydroxyindole and stimulated by isatin. Appl. Environ. Microbiol. 2007, 73, 4100–4109.
- Chakraborty, S.; Deokule, J.S.; Garg, P.; Bhattacharya, S.K.; Nandy, R.K.; Nair, G.B.; Yamasaki, S.; Takeda, Y.; Ramamurthy, T. Concomitant infection of enterotoxigenic Escherichia coli in an outbreak of cholera caused by Vibrio cholerae O1 and O139 in Ahmedabad, India. J. Clin. Microbiol. 2001, 39, 3241–3246.
- Holm, A.; Vikström, E. Quorum sensing communication between bacteria and human cells: Signals, targets, and functions. Front. Plant Sci. 2014, 5, 309.
- Doherty, N.C.; Tobias, A.; Watson, S.; Atherton, J.C. The Effect of the Human Gut-Signalling Hormone, Norepinephrine, on the Growth of the Gastric Pathogen Helicobacter pylori. Helicobacter 2009, 14, 223–230.
- Nakano, M.; Takahashi, A.; Sakai, Y.; Kawano, M.; Harada, N.; Mawatari, K.; Nakaya, Y. Catecholamine-induced stimulation of growth in Vibrio species. Lett. Appl. Microbiol. 2007, 44, 649–653.
- Belay, T.; Sonnenfeld, G. Differential effects of catecholamines on in vitro growth of pathogenic bacteria. Life Sci. 2002, 71, 447–456.
- Yang, Q.; Anh, N.D.Q.; Bossier, P.; Defoirdt, T. Norepinephrine and dopamine increase motility, biofilm formation, and virulence of Vibrio harveyi. Front. Microbiol. 2014, 5, 584.
- Hegde, M.; Wood, T.K.; Jayaraman, A. The neuroendocrine hormone norepinephrine increases Pseudomonas aeruginosa PA14 virulence through the las quorum-sensing pathway. Appl. Microbiol. Biotechnol. 2009, 84, 763–776.
- Stevens, E.A.; Mezrich, J.D.; Bradfield, C.A. The aryl hydrocarbon receptor: A perspective on potential roles in the immune system. Immunology 2009, 127, 299–311.
- Fujiya, M.; Musch, M.W.; Nakagawa, Y.; Hu, S.; Alverdy, J.; Kohgo, Y.; Schneewind, O.; Jabri, B.; Chang, E.B. Short Article The Bacillus subtilis Quorum-Sensing Molecule CSF Contributes to Intestinal Homeostasis via OCTN2, a Host Cell Membrane Transporter. Cell Host Microbe 2007, 2, 299–308.
- Hong, H.A.; Khaneja, R.; Tam, N.M.K.; Cazzato, A.; Tan, S.; Urdaci, M.; Brisson, A.; Gasbarrini, A.; Barnes, I.; Cutting, S.M. Bacillus subtilis isolated from the human gastrointestinal tract. Res. Microbiol. 2009, 160, 134–143.
- Tam, N.K.M.; Uyen, N.Q.; Hong, H.A.; Duc, L.H.; Hoa, T.T.; Serra, C.R.; Henriques, A.O.; Cutting, S.M. The Intestinal Life Cycle of Bacillus subtilis and Close Relatives. J. Bacteriol. 2006, 188, 2692–2700.
