Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
N6-metyladenosine (m6A), one of the most common RNA methylation modifications in mammals, has attracted extensive attentions owing to its regulatory roles in a variety of physiological and pathological processes. As a reversible epigenetic modification on RNAs, m6A is dynamically mediated by the functional interplay among the regulatory proteins of methyltransferases, demethylases and methyl-binding proteins. It has become increasingly clear that m6A modification is associated with the production and function of microRNAs (miRNAs).
- N6-metyladenosine
- methyltransferases
- demethylases
- methyl-binding proteins
- miRNAs
1. Introduction
N6-metyladenosine (m6A) modification refers to the methylation that occurs at the 6th nitrogen atom of adenine. Since firstly found in 1974, m6A has received extensive attention for regulating abundant internal modifications on various RNAs, including mRNAs, microRNAs (miRNAs), small nuclear RNAs (snRNAs), long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs) [1,2,3]. As we all know, the formation of m6A modification is dynamically catalyzed by methyltransferases and demethylases, also called “writers” and “erasers”, respectively. The binding proteins, named “readers”, can specifically combine with m6A modification and mediate m6A biological function in different pathological and physiological processes [4]. At present, m6A modification has been widely recognized as a reversible and dynamic epigenetic modification on various RNAs [5], and it is becoming increasingly clear that m6A potentially contributes to the occurrence and development of multiple diseases through altering RNA expressions or RNA functions [6]. Recently, the clinical value of m6A modification in diseases has become apparent, and m6A modification has been commonly utilized as a promising biomarker to diagnose, prevent and treat diseases [7].
As the most common non-coding RNAs, miRNAs exert their biological functions through interaction with RNAs or proteins [8]. Recently, emerging works of literature have demonstrated that miRNAs were modified by various chemical modifications, such as m6A, m1C, m5C and m7G, which affect the processing and functions of corresponding miRNAs [9,10,11]. Among these modifications, m6A attracted the most attention. As reported previous study, m6A modification is preferentially concentrated near the 3′-UTR, of which 67% contain ncRNAs, such as the binding sites of miRNAs, implicating that m6A and ncRNAs may jointly regulate target mRNAs through cooperation or competition [12]. The latest studies have found that m6A modification affects the cleavage, transport, stability and degradation of corresponding miRNAs and influences the interactions between miRNAs and long non-coding RNAs or proteins [13,14,15]. Interestingly, it is becoming increasingly clear that miRNAs also have critical roles in regulating m6A modification by changing the regulatory proteins of m6A modification [16].
2. Dynamic Regulation of m6A Modification
In recent years, more and more studies have found that m6A modification on RNAs can be dynamically regulated by dedicated methyltransferases, demethylases or methyl-binding proteins. A set of these regulatory proteins have been summarized in Figure 1.
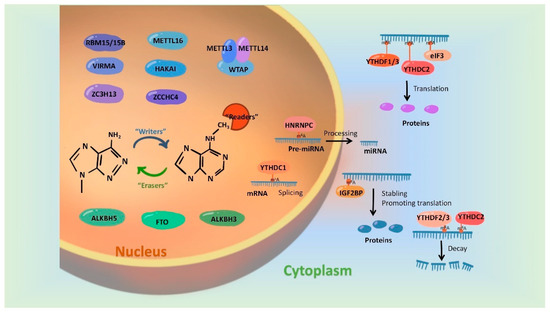
Figure 1. Regulatory proteins involved in mediating the m6A modification. m6A modification is synergistically catalyzed by methyltransferases (Writers), demethylases (Erasers) and methyl-binding proteins (Readers). The formation of m6A was initially catalyzed by a group of complexes, in which METTL3 and METTL 14 act as the active center of methyltransferases and WTAP, RBM15/15B, VIRMA and ZC3H13 play the part of adaptor proteins. In addition, METL16, HAKAI and ZCCHC4 were evidenced to be independently catalyzed the formation of m6A modification. FTO, ALKBH5 and ALKBH3 have been identified as m6A demethylases and they remove m6A marks on RNAs in an independent manner. Readers, involving biological functions by recognizing m6A modifications, include the YTH family, HNRNP family, IGFBPP1/2/3, EIF3 and ABCF1.
3. Mutual Regulatory Mechanisms between m6A Modification and miRNAs
Recently, a large number of studies have demonstrated that m6A modification on miRNAs play essential roles in various pathophysiological processes. Most interestingly, emerging evidence revealed that miRNAs also regulate m6A modification by altering expressions of m6A regulatory proteins [51,55]. So far, mutual regulatory mechanisms between m6A modification and miRNAs in multiple diseases have attracted a huge amount of attention.
As a series of noncoding and single-stranded small molecular RNA with a length of 18–24 nucleotides, miRNAs could target specific mRNA sites and promote degradation or inhibit translation of mRNA [56]. Although there are nearly 3000 miRNAs in mammals, the generation processes of different miRNAs are almost consistent. In detail, upon transcription from DNA, primary transcripts of miRNAs (pri-miRNAs) are spliced by RNase Ⅲ structure domain proteins and double-stranded RNA binding protein Drosha and Di George Syndrome critical region 8 (DGCR8) to form precursor of miRNAs (pre-miRNAs) in the nucleus. Then, pre-miRNAs are exported from nucleus into cytoplasm by forming a complex with a transporter protein exportin-5 and a GTP-binding nuclear protein Ran-GTP [53,54]. Once transported out of nucleus, pre-miRNAs are cleaved into mature miRNAs through another type III RNase Dicer. The mature miRNAs subsequently bind to mRNAs with the help of Ago proteins, thereby affecting the levels or translation processes of corresponding mRNAs [57]. Interestingly, advances in m6A modification in recent years have widely broadened mechanisms underlying miRNA processing and regulation. Specifically, emerging studies have shown that m6A modification and its regulatory proteins involve in the production of mature miRNAs, which in turn affect the level of m6A modification [58].
3.1. m6A Modifications Involves in miRNA Generation and Function
Current studies have indicated that m6A modification is involved in the generation process of miRNAs, thus affecting the level of mature miRNAs. Published results in the journal Nature in 2015 revealed that decreasing the level of m6A modification on pri-miRNAs by knocking down METTL3 expression could inhibit the binding of DGCR8 to pri-miRNAs, which led to about 70% miRNAs being downregulated by at least 30% [57]. Up to now, the mechanism according to which reduction of m6A modification on pri-miRNAs inhibits the maturation of miRNAs in a DGCR8-dependent manner has been found in different diseases. For example, catalyzed by over-expressed METTL3, high m6A modification can promote the maturity of miR-25 and miR-25-3p by strengthening the combination of DGCR8 and pri-miR-25 in pancreatic duct epithelial cells, and this may provoke malignant phenotype of pancreatic cancer cells [59]. The reduction level of m6A modification mediated by low expression of METTL3 and METTL14 makes the weaker recognition of pri-miR-126 by DGCR8, which hinders the maturation of miR-126, thereby activating the PI3K/AKT/mTOR pathway to promote the proliferation and activation of fibroblasts. Moreover, METTL3-dependent m6A was involved in the DGCR8-mediated maturation of pri-miR-126 in endometriosis development [60]. In addition, the interaction of METTL3 and DGCR8 positively modulates the biogenesis process of miR-873-5p, miR-365-3p and miR-221/222 in an m6A-dependent manner in different pathological processes, and as the simplest for specific miRNA, miR-873-5p participated in fighting colistin induced oxidative stress and apoptosis in kidney injury [61], miR-365-3p regulated chronic inflammatory pain induced by Complete Freund’s Adjuvant in the spinal cord [62], and miR-221/222 negatively mediate the PTEN expression, thus leading to the proliferation of bladder cancer cells [63]. In addition, cigarette smoke can stimulate the production of excess mature miRNA-93 in bronchial epithelial cells via enhanced m6A modification, which was mediated by overexpressed METTL3 [64]. METTL3 also plays a major catalytic role in m6A modification in unilateral ureteral obstruction mice and drove obstructive renal fibrosis development by promoting miR-21-5p maturation [65]. In addition, it has been indicated that silencing of METTL3 expression can elevate the levels of pri-miR-663 and m6A methylation-modified pri-miR-663, which resulted in suppressing of miR-663 maturation process in A549 and PC9LC cells [66]. In a manner similar to METTL3, the METTL14-mediated m6A marks also enhanced the recognition of pri-miR-126 by DGCR8, thus subsequently processing to mature miRNA-126, which is involved in hepatocellular carcinoma metastasis [60]. Different from the above mechanisms, METTL3 induced upregulation of miR-143-3p mostly depends on the shear effect of Dicer on pre-miR-143-3p in lung cancer cells [67]. Moreover, as in bone marrow-derived mesenchymal stem cells, METTL3 also methylate pre-miR-320, on which m6A modification is a key factor that is recognized and decayed by YTHDF2 [68]. METTL3 promoted the transition from pri-miR-1246 to mature miR-1246, of which upregulation can significantly enhance the metastasis ability of colorectal cancer cells [69]. METTL3-mediated m6A modification also promotes the expressions of 9 miRNAs, including miR-106b, miR-18a, miR-18b, miR-3607, miR-432, miR-30a, miR-320b, miR-320d and miR-320e, and bioinformatics analysis has shown that these miRNAs are involved in regulating signaling pathways closely related to malignant transformation induced by arsenite [70]. In addition, four miRNAs (miR-130a-3p, miR-130b-3p, miR-106b-5p and miR-301a-3p) are all related to short overall survival of kidney renal clear cell carcinoma patients and have significantly negative correlation with METTL14 mRNA [71]. Up to now, the importance of methyltransferases-catalyzed m6A modification on pri-miRNAs has been widely recognized, and a variety of methyltransferase components can affect the generation and function of miRNAs.
Besides methyltransferases, m6A demethylases and methyl-binding proteins are also involved in miRNA biogenesis. The earliest study showed that knocking down the FTO expression significantly increased levels of 42 miRNAs and decreased levels of 9 miRNAs [72]. A subsequent study reported that FTO regulates cell migration and invasion in breast cancer cells by inhibiting miR-181b-3p [73]. Moreover, FTO has been well evidenced to promoted bladder cancer cell proliferation via the FTO/miR-576/CDK6 pathways [74]. ALKBH5 inhibits tumor growth and metastasis by inhibiting miR-107/LATS2 mediated YAP activity in non-small cell lung cancer [75]. Peng et al. indicated that ALKBH5, the most potent member related to patient outcomes and to suppressing esophageal cancer malignancy in cell and animal models, demethylated pri-miR-194-2 and inhibited miR-194-2 biogenesis through an m6A/DGCR8-dependent manner [76]. Interestingly, in human non-small cell lung cancer cells, the depletion of ALKBH5 did not change the miR-21-5p level but altered the m6A abundance on miR-21-5p, thereby changing the miR-21-5p silencing potency towards its target mRNAs, which finally impaired the proliferation and motility of human non-small cell lung cancer cells [77]. In addition, ALKBH5 demethylated pri-miR-320a-3p, thus blocking DGCR8 from interacting with pri-miR-320a-3p and leading to mature process blockage of pri-miR-320a-3p in silica-inhaled mouse lung tissues [78].
In addition to being directly affected by m6A methyltransferases and demethylases, miRNA generation is also regulated by m6A methyl binding proteins. YTHDC1, a well-known m6A reader, facilitated the biogenesis of mature miR-30d via m6A-mediated regulation of mRNA stability. Furthermore, miR-30d represses pancreatic tumor genesis via suppressing aerobic glycolysis [79]. m6A reader protein HNRNPA2B1 also binds to a subset of m6A-modified pri-miRNA transcripts, thus interacting with DGCR8 and promoting primary miRNA processing, and depletion of HNRNPA2B1 caused a reduction in the levels of 61 miRNAs in HEK293 cells. Moreover, transiently overexpressed (5.4-fold) HNRNPA2B1 in MCF-7 cells led to significant alteration of more than 100 miRNAs, which regulate TGFβ and Notch signaling pathways according to MetaCore Enrichment analysis [80,81,82]. Yi et al. have reported that miR-185 transfer from vascular smooth muscle cells to endothelial cells is controlled by HNRNPA2B1 [83], but the role of m6A modification in this mediate process needs to be further explored. In addition, HNRNPA2B1 reads the m6A site on pri-miR-106b or pri-Let-7b to facilitate the maturing of miR-106b-5p or Let-7b in the lung cancer cells [84,85]. Another m6A binding protein, IGF2BP1, promotes serum response factor expression in an m6A-dependent manner by impairing the miRNA-directed downregulation of the serum response factor (SRF) mRNA in cancer cells [86]. In addition to regulating the generation process of miRNAs, m6A modification can directly modify mature miRNAs and affect their stability and degradation [72]. Of note, m6A modification on the E2F transcription factor 3 (E2F3) mRNA was required for the interaction between miR-660 and E2F3 mRNA in gastric cancer, indicating that m6A also affects the function of miRNA apart from participating in its production process [87].
In a word, emerging studies have identified the roles of m6A modification during the processing and maturation of miRNAs, which will surely provide good candidate targets for miRNA intervention. Although mechanisms underlying m6A modification affecting miRNA generation and function are diverse and complex, the general mechanisms are similar. We drew a schematic diagram, which take miR-30d [79], miR-21-5p [65] and pre-miR-25 [59] as examples, to show the specific mechanisms of m6A regulatory proteins regulating miRNAs (Figure 2).
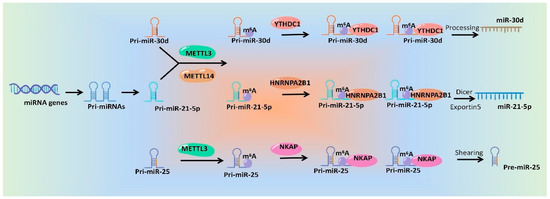
Figure 2. Schematic diagram of the mechanisms of m6A modification and its regulatory proteins regulating miRNA production.
3.2. miRNAs Regulate the m6A Modification
Since miRNAs affect the protein level through interacting with mRNAs and m6A modification is a dynamic reversible methylation [88], it is rationality that miRNAs are involved in the regulation of m6A modification by affecting the regulatory proteins. At present, several studies have shown that miRNAs regulate m6A modification via sequence pairing of mRNAs of methyltransferases, demethylases, and methyl-binding proteins in various tissues [89]. In detail, METTL3 was identified as the direct target of miR-1269b [90] and miR-338-5p [91], thus inhibiting gastric cancer development. miR-33a is capable of reducing the METTL3 expression at both mRNA and protein levels, thus affecting proliferation, survival and invasion of non-small cell lung cancer [92]. Moreover, miR-600 can attenuate METTL3 expression and restrain the migration and proliferation of lung cancer cells [93]. Similarly, the down regulation of miR-524-5p also up-regulates the expression of METTL3 in non-small cell lung cancer cells [94]. miR-4429 targeted and repressed METTL3 to inhibit m6A-mediated stabilization of SEC62, a component belonging to tetrameric Sec62/Sec63-subcomplex of Sec-complex, thus hindering proliferation and encouraging apoptosis in gastric cancer cells [95]. Moreover, Cai et al. concluded that mammalian hepatitis B X-interacting protein (HBXIP) suppresses miRNA let-7g, thus up-regulating METTL3, which in turn promotes the expression of HBXIP through m6A modification, leading to stimulation or proliferation of breast cancer cells [96]. As an independent prognostic factor in hepatoblastoma patients, METTL3 was identified as a direct target of miR-186, of which low level led to high expression of METTL3, thus significantly inhibiting the proliferation, migration and invasion of hepatoblastoma cells [97]. Moreover, miR-320d has been evidenced to target METTL3, thus affecting KIF3C expression through changing m6A modification on KIF3C mRNA in prostate cancer cells [98]. Under the treatment of Mono-(2-ethylhexyl)phthalate (MEHP), miRNAs such as miR-16-1-3p, miR-101a-3p, miR-362-5p, miR-501-5p, miR-532-3p and miR-542-3p are dramatically activated in murine macrophage Raw 264.7 cells, and these miRNAs are all predicted to regulate METTL14, thus promoting m6A modification in Scavenger Receptor B type 1 (SR-B1) mRNA [99]. Cui et al. reported that miR-193a-3p directly targets ALKBH5 to inhibit the growth and promote the apoptosis of glioma cells by suppressing the AKT2 pathway both in vitro and in vivo [100]. Interestingly, circGPR137B acted as a sponge for miR-4739 to up-regulate its target FTO, which mediated m6A demethylation of circGPR137B and promoted its expression, thus finally forming a feedback loop comprising circGPR137B/miR-4739/FTO axis and affecting the hepatocellular carcinoma cells [101]. Results from Yang et al. indicated that imiR-155 directly targets FTO to negatively regulate its expression and increase m6A level in renal clear cell carcinoma cells. Regarding specific mechanisms, miR-155 is directly bound to the 3′-UTR of FTO mRNA and reduced FTO protein levels [102].
The methyl binding proteins of m6A modification are also directly targeted by miRNAs. In detail, Zheng et al. reported that miRNA-421-3p targets YTHDF1 to inhibit p65 mRNA translation, thus preventing inflammatory response in cerebral ischemia/reperfusion injury [103]. miR-376c also has been indicated to negatively modulate YTHDF1 expression in non-small cell lung cancer cells [104]. Negative correlations between the miR-145 level and YTHDF2 mRNA expression were observed in hepatocellular carcinoma [105] and epithelial ovarian cancer cells [106], and further detecting results showed that miR-145 decreased the luciferase activities of 3′-UTR of YTHDF2 mRNA, implicating that YTHDF2 is the direct target gene of miR-145 [105,106]. In addition, YTHDF2 mRNA is also regulated by miRNA-495 in prostate cancer cells [107] and miR-6125 in colorectal cancer cells [108]. Bioinformatics analysis from Hao et al. literature revealed IGF2BP1 as the putative target of miR-670, of which mimics and inhibitors were microinjected into parthenogenetic activation embryos, thus confirming these findings [109]. IGF2BP2, another m6A methyl binding protein, is highly expressed in thyroid cancer cells and identified as a target of miR-204 [110]. In addition, inhibition of miR-133b also resulted in the up regulation of IGF2BP2 in colorectal cancer cells [111]. Different from the above-mentioned mechanisms where miRNAs regulated m6A modification, results from Chen et al. indicated that overexpressing dicer increased the m6A modification level, and this was not achieved by alternating the quantity of m6A methyltransferases or demethylases in mouse embryonic fibroblasts. Further experiments showed that miRNAs regulate activity and location of METTL3, which subsequently modulate m6A modification and impede the reprogramming of mouse embryonic fibroblasts to pluripotent stem cells [112].
In a word, miRNAs can influence m6A modification by regulating the regulatory proteins of m6A and ultimately participate in a variety of pathological and physiological processes. We drew a schematic diagram, in which we take METTL3 [93], FTO [113] and YTHDF2 [108] as examples, to show the miRNAs involving in regulation of m6A modification and its biological effect (Figure 3).
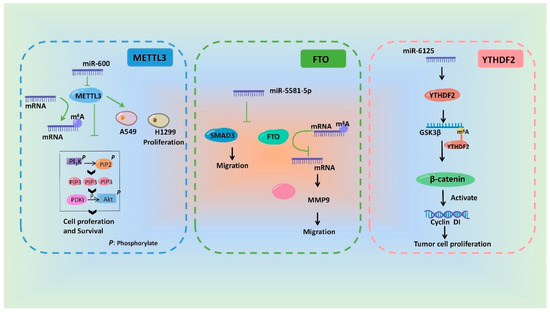
Figure 3. Schematic diagram of the mechanisms showing that m6A modification and its biological effects are regulated by miRNAs.
As mentioned above, m6A modification is regulated by different m6A regulatory proteins in a variety of diseases by promoting biosynthesis of miRNAs, and miRNA regulates the biological functions of m6A regulatory proteins. Based on this interplay, we summarized the change trends and regulation relationships of m6A regulatory proteins and miRNAs in tissues or cells during the occurrence and development of different diseases, as shown in Figure 4.
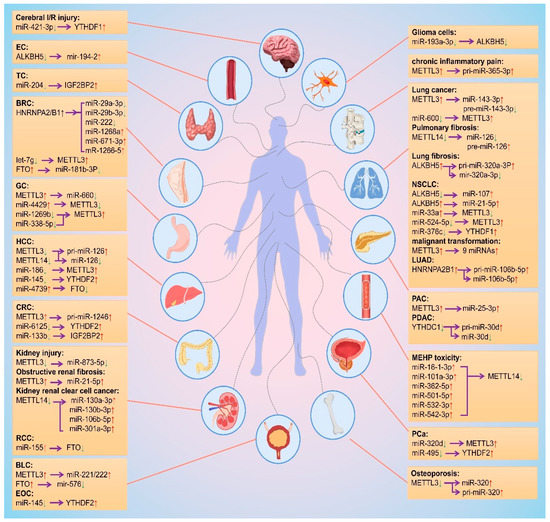
Figure 4. Mutual regulation between m6A modifications and miRNAs. The red and green arrows represent the level rise and fall, respectively. The purple arrows point to the regulated object. EC: esophageal cancer; TC: thyroid cancer; BRC: breast cancer; GC: gastric cancer; HCC: hepatocellular carcinoma; CRC: colorectal cancer; RCC: Renal cell carcinoma; BLC: bladder cancer; EOC: epithelial ovarian cancer; NSCLC: non-small cell lung cancer; LUAD: lung adenocarcinoma; PAC: pancreatic cancer; PDAC: Pancreatic ductal adenocarcinoma; PCa: prostate cancer.
This entry is adapted from the peer-reviewed paper 10.3390/ijms24010773
This entry is offline, you can click here to edit this entry!
