This entry mainly gives an overview of the structure of the Ccr4-Not complex, its major components and their enzymatic activities. In the accompanying manuscript the biological roles of the complex is discussed in detail as well as clinical conditions associa
- Ccr4–Not complex
- mRNA decay
- deadenylation
- transcription
- cell cycle
- chromatin modification
- RNA export
- DNA damage repair
- Human Disease
The mammalian Ccr4–Not complex, carbon catabolite repression 4 (Ccr4)-negative on TATA-less (Not), is a large, highly conserved, multifunctional assembly of proteins that acts at different cellular levels to regulate gene expression. (Draft for Definition)
IntroductionThe yeast Ccr4–Not complex is a large (1.9-MDa) and highly conserved multifunctional assembly of proteins, involved in different aspects of mRNA metabolism. These include the repression and activation of transcription initiation, control of mRNA elongation, deadenylation-dependent mRNA turnover. The complex is also involved in ubiquitin-protein transferase activity reviewed [1–5]. The activation of the complex can be seen in various ‘downstream effects’ such as histone methylation and cell cycle regulation. Most of the original studies concentrated on the role of the Ccr4–Not complex in Saccharomyces cerevisiae. In yeast the complex has nine core subunits, comprising Ccr4 (carbon catabolite repression), Caf proteins (Ccr4 associated factor) (Caf1, Caf40, Caf130) and Not proteins (Not1, Not2, Not3, Not4, and Not5) as well as several less strongly associated components which are probably interacting partners [6,7] (Table 1). Not1 is the largest subunit of the complex ( >200 kD) and forms a scaffold for the other components [8]. No clear function has been assigned to the Not module comprising Not2, Not3 and Not5 although it does act as a cofactor for the deadenylation activity and may be involved in interaction with the ribosome [9,10]. Genetic approaches in yeast have also demonstrated that the Not1–4 genes can globally repress RNA polymerase II activity. The mutation of these genes increases the basal expression of many genes [11]. The Ccr4-Caf1 mRNA deadenylase complex contains a 3′ exoribonuclease which is involved in removing poly (A) tails from mRNA [12–16]. Not4 is responsible for the second major enzymatic activity of the complex, E3 ligase-mediated ubiquitylation [17].
Complexes of a comparable size, containing the human orthologues CNOT1–CNOT9 with three additional subunits of CNOT10, Tab182 (Tankyrase 1-binding protein1, TNKS1BP1) and C2ORF29 (CNOT11), have been identified in mammals (Table 1). Four deadenylase subunits are expressed in mammalian cells. These appear to form various heterodimers, CNOT7/CNOT6, CNOT7/CNOT6L, CNOT8/CNOT6, and CNOT8/CNOT6L. Thus, the complex contains either CNOT7 or CNOT8, suggesting they compete for binding to CNOT1 [18,19]. While the E3 ubiquitin ligase Not4 is consistently present in the yeast complex, CNOT4 is not as stably associated as the other subunits in mammalian cells [18]. No orthologues of CNOT10, CNOT11, or Tab182 have been identified in yeast [20]. The CNOT3 subunit, with no specific enzymatic activity, is orthologous to two yeast subunits, Not3 and Not5. Every individual subunit appears to have a unique role with a slight overlap between some proteins [21]. The evidence in support of this includes the observation that mutations and deletions of each different subunit are responsible for different phenotypes in yeast [21].
Structure of the Ccr4–Not (CNOT) Complex
As with any large multi-subunit assembly of proteins considerable effort has been expended to determine the structure of Ccr4–Not. Up to the present, detailed structural information is available for the complex from four species: Schizosaccharomyces pombe, Saccharomyces cerevisiae, Drosophila melanogaster, and Homo sapiens. Limited data are also available for the thermophilic fungus Chaetomium thermophilum complex [9,14,16,22–27]. In all cases, it has been shown that CNOT1, the largest subunit ( > 200 kD molecular mass), forms a scaffold for the complex and most, although not all, other components bind to it directly [28]. However, a number of obvious differences between the species have been identified. Firstly, there are four deadenylase components in the human complex (CNOTs 6, 6L, 7 and 8) but only two in yeast (Ccr4 and Caf1). The Ccr4-Caf1 mRNA deadenylase complex contains a 3′ exoribonuclease motif [12–16]. Secondly, it appears that the E3 ligase protein, CNOT4/Not4, is quite strongly associated with the complex in yeast but not in Drosophila or humans[17,18,29]. Thirdly, additional components have been identified in the Drosophila and human complexes. These are CNOT10 and CNOT11 (C2orf29). TNKS1BP1 (Tab182) also seems to be a member of the human complex [18,20,30] (Table 1).
Table 1. A list of alternative names for equivalent CCR4–NOT complex subunits from Saccharomyces cerevisiae (baker’s yeast), Homo sapiens (human) and Drosophila melanogaster (fruit fly).
|
Saccharomyces Cerevisiae
|
Homo Sapiens
|
Drosophila Melanogaster
|
Function
|
|
NOT1/CDC39 |
CNOT1 |
NOT1 |
Scaffold |
|
NOT2/CDC36 |
CNOT2 |
ReginaNOT2 |
Unknown but contributes to stabilization of the complex and RNA substrate recruitment |
|
NOT3 |
Not present |
Not present |
Unknown but contributes to stabilization of the complex and RNA substrate recruitment |
|
NOT5 |
CNOT3 |
NOT3 |
Interaction with ribosomes |
|
NOT4/MOT2/SIG1 |
CNOT4 |
NOT4 |
Ubiquitin E3-ligase activity |
|
CCR4 |
CNOT6, CNOT6L |
Twin, CCR4 |
Deadenylase |
|
CAF1 |
CNOT7, CNOT8 |
P0P2, CAF1 |
Deadenylase |
|
CAF40 |
CNOT9 (RQCD1) |
RCD1 |
Transcriptional cofactor
|
|
Not present |
CNOT10 |
NOT10 |
Unknown but contributes to stabilization of the complex and RNA substrate recruitment |
|
Not present |
CNOT11 (C2orf29) |
NOT11 |
Unknown but contributes to stabilization of the complex and RNA substrate recruitment |
|
CAF130 |
Not present |
Not present |
Unknown |
|
Not present |
TNKS1BP1 (Tab182) |
Not present |
Multifunctional |
The complex, of approximately 1.9M molecular weight in yeast, is L-shaped with two arms of approximately equal length (180–190Å) and a hinged region present in the centre [22,24].
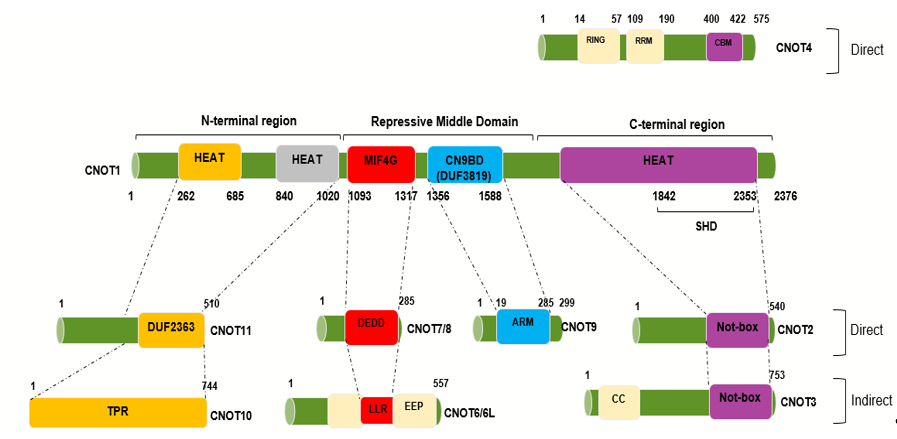
The complex is assembled on the NOT1 backbone. How the other components associate has been determined using in vitro protein binding studies, crystallography, and electron microscopy. Binding sites for the other components of the complex on CNOT1 appear to consist of α-helical domains. A number of structural sub-complexes have been delineated-these comprise the deadenylase module (Ccr4 and Caf1 in yeast, CNOT6/CNOT6L and CNOT7/CNOT8 in humans), the NOT4 (Not4) the E3 ligase module and the ‘Not module’ (Not2 and Not5 in yeast, CNOT2 and CNOT3 in humans). The binding sites of additional subunits have also been mapped-thus, metazoan CNOT10 and CNOT11 bind to the N-terminal domain of CNOT1 and CNOT9 (Caf40 in yeast) binds to the central region of CNOT1 [16,20,25,31] (Figure 1). The C-terminal region of CNOT1 forms a rigid structure, which comprises two perpendicular stacks of HEAT-like repeats. CNOT2 and CNOT3 each contain SH3-like Not box domains which provide dimerization sites. CNOT2/CNOT3 binds to the CNOT1 C-terminal region [23,28] (Figure 1). The C-terminal binding site of CNOT1 comprises 10 HEAT repeats which contain helix A-turn-helix B motifs. The structures of the C-terminal complexes have been determined for the yeast and human proteins. CNOT2/Not2 and CNOT3/Not5 form a heterodimer through their Not-box motifs. A region of CNOT3/Not5 interacts with HEAT repeats 1–5 through hydrophobic and polar amino acids and a region of CNOT2/Not2 is spread across Not1 from HEAT repeats 9 and 10 and binding to repeats 4–6 [9,23]. Although no specific functions have been determined for this C-terminal module it is linked to the stability of the complex as a whole and the recruitment of mRNAs [9]. The C-terminal complex associates with synthetic ribonucleotides, such as poly(U) RNA, in vitro with a binding site comprising structural elements from Not1, Not2 and Not5 [9]. Boland et al. have also shown that an intact C-terminal module of the Drosophila CCR4–NOT complex is required for optimal mRNA degradation [23]. Furthermore, the CNOT2/CNOT3 heterodimer can stimulate the deadenylase activity of the complex [23]. Recent evidence indicates that yeast Not5 directly associates with the ribosome and, together with Not4, plays an important role in the regulation of mRNA half-life (Section 3) [10].
CNOT4 (Not4) is an evolutionarily conserved E3 ubiquitin ligase [17,29]. It contains a RING domain, a linker region which tends to form a coiled-coil, an RNA recognition motif (RRM) domain, and a C3H1-type zinc finger domain (ZNF) [31]. These motifs are all present within the conserved N-terminal region of human CNOT4. The C-terminal region of metazoan Not4 is variable in sequence but contains a conserved Caf binding motif (CBM) through which it binds directly to Caf40 (CNOT9) [31]. Flanking sequences to the CBM are involved in interaction with Not1 as well as assisting with Caf40 binding. This motif is not present in the yeast Not4 proteins although a Not1 binding site has been described [9]. It is not fully clear why yeast Not4 associates strongly with the full complex whereas the human and Drosophila proteins do not. However, it has been shown that the C-terminal region of human CNOT4 (Not4-C) is able to bind to the complex [31]. It has been suggested that the N-terminal region, therefore, somehow prevents Not4-C from interacting in the human complex. This could be explained by possible post-translational modifications or additional binding partners [31]. As well as binding to Caf40/CNOT9 the Not4-C also interacts with the C-terminal HEAT region of CNOT1, with a relatively slight contribution from CNOT2 and CNOT3 [9,31] (Figure 1). A number of substrates for ubiquitylation by CNOT4 have been identified and these are discussed elsewhere in this review (Section 4). Significantly, CNOT4 is required for optimal deadenylation activity by the full human CNOT complex, although a short C-terminal peptide will substitute for the whole protein [31].
The CNOT9 (Caf40) subunit binds to a central CNOT1 coiled coil domain (termed CN9BD or DUF3819). This is adjacent to the CNOT1 MIF-4G (middle portion of eIF4G) region (Figure 1). The human CNOT9 monomer contains six armadillo repeats forming a solvent-accessible, positively-charged cleft 21–22 Å wide [32]. Armadillo repeats are normally involved in protein-protein interactions but it has been shown that CNOT9 can also bind certain oligonucleotides in in vitro assays [32].
The deadenylase module associates with the central HEAT repeat-containing MIF-4G region of Not1. Thus, Caf1 (CNOT7) binds to Not1 through pre-existing structural motifs, made up of conserved hydrophobic residues. This allows full access for RNA molecules to the active site on Caf1 [15,16]. The leucine rich repeat (LRR) domain of Ccr4 (CNOT6L) binds to Caf1. Caf1 interacts with Ccr4 via a surface formed by a long loop and an α-helix. This region of Caf1 undergoes a localized conformational change compared to the unbound structure [16]. In humans, two additional deadenylase components are present in the complex, CNOT6 and CNOT8, and it seems likely that they bind to CNOT1 in a comparable way to CNOT6L and CNOT7 (Figure 1).
One of the obvious differences between the yeast and metazoan CNOT complexes is the presence of CNOT10 and CNOT11 (C2orf29) in the latter complex. Human and Drosophila CNOT11 binds to the N-terminal region of CNOT1 and CNOT10 binds to it [20]. The area of Not1 between the MIF-4G region and the N-terminal region comprises thirteen HEAT domains. The Not1 a.a.154–753 structure has been shown to contain antiparallel helices assembled side by side to form an elongated molecule. It has been suggested that these form structures ideal for protein–protein interactions within the complex or for binding of other molecules [16]. It should be noted, however, that this structure was determined for yeast CNOT1 which has no CNOT10 and 11 orthologs. Although CNOT10 and 11 do not appear to have any enzymatic activity of their own, their presence stimulates deadenylation through stabilization of the RNA substrate. It has been suggested that Caf40 (CNOT9) is the major enhancer of deadenylation, probably due to its proximity to the exonucleases, but if this is not available, CNOT10 and CNOT11 can compensate [25].
Figure 1. Human CNOT1 interaction regions with the other CNOT subunits. Interaction map obtained from negative-stain electron microscopy showing the interaction sites between CNOT1, acting as a scaffolding platform, and the other CNOT subunits. The human homologue, CNOT1, encompasses 2376 amino acids and is 20% identical (32% similar) to its yeast counterpart. Different colors represent the different regions as shown above. The human Tab182 interacting site has not been mapped yet. Negative on TATA-less (NOT), Middle domain of eukaryotic initiation factor 4G (MIF4G), Domain of uncharacterized function (DUF3819 and DUF2363), Exonuclease-endonuclease-phosphatase (EEP)-DNase I-like domains, Death effector domain-containing protein (DEDD)-RNase D-like domains, Leucine-rich region (LRR)-RNase D-like domains, Superfamily Homology Domain (SHD), Armadillo (ARM) Repeat Domain, predicted coiled-coil domain (CC), Predicted domain consisting of α-helical TPR-like repeats (TPR), CAF40-binding motif (CBM) (Adapted from [25,31,33,34]).
Depletion of CNOT1 results in the destabilization of the whole complex and degradation of some other subunits such as CNOT2, CNOT6L, CNOT7, and CNOT9, but not CNOT3, in HeLa cells [35]. Although, CNOT1 has no enzymatic activity, as far as is known, the importance of its scaffolding function for the deadenylase activities of the CNOT complex cannot be overstated. It has been shown that in CNOT1-depleted HeLa cells the level of CHOP mRNA increased and the cells undergo caspase-4 activation causing ER stress-mediated apoptosis, indicating CNOT1 is essential for viability and cell proliferation [35]. In addition, CNOT1 depletion in HeLa cells reduces the deadenylase activities and decreases the level of P-body formation, where mRNA decay is thought to take place [35].
What are considered to be the major constituents of the CNOT complex are described above. However, a number of other proteins have been routinely found to associate with the complex [6,7,36]. It appears that these components are not integral but are required for routine functions. For example, the BTG/Tob complex binds to CNOT7 and has a role in the recruitment of mRNAs [37]. Consequences of the interaction have been reported variously to be either activation or repression of deadenylase activity [38–40]. A large number of other RNA binding proteins (for example, Nanos2, Pumilio, Smaug and Tristetrapolin) act as adaptor proteins and interact with the CNOT complex and cause the suppression of target mRNAs [36]. Recent evidence suggests that Puf3 and Zfs1 associate with the complex and are critical in mRNA substrate selectivity as well as enhancing RNA binding [41]. Puf3 can distinguish between RNAs of very similar sequence and can therefore facilitate the ability of the Ccr4–Not complex to regulate the level of expression of particular target proteins [41]. Interaction of the complex with GW182 (a component of miRISC) through CNOT1 plays a role in miRNA-induced deadenylation [42–44]. The core subunit, CNOT1, has also been shown to associate with the translational repressor and decapping activator, the DEAD box protein DDX6 [34,45–47] (Section 3). DDX6 plays a role as a translational repressor in different pathways, including mRNA storage in erythropoiesis and microRNA mediated gene silencing [47]. Other proteins shown to interact with the CNOT complex include Bag-of-marbles, which represses the expression of specific mRNAs and binds to CAF40 (CNOT9), HIPK family kinases, which associate with the CCR4–NOT components CNOT2 and CNOT3 and phosphorylate the complex, the transcription factor EBF1, which binds CNOT3 and yTAFI, a core component of TFIID, which binds Not1 [48–51]. A number of these are discussed in more detail in the following sections.
Interestingly, it has been shown that the Ccr4–Not complex strongly associates with the YTH domain of nucleus-specific RNA binding subunit, Mmi1 (meiotic mRNA interceptor 1), close to the nuclease module, in Schizosaccharomyces pombe [24]. In fission yeast, Mmi1 is essential for viability. It represses the expression of transcripts and increases the deadenylation of target RNAs in in vitro assays [8].
In the accompanying manuscript, the role of the Ccr4-Not complex in various biological processes is discussed in detail as well as its links to various clinical conditions.
References
- Denis CL, Chen J. The CCR4-NOT complex plays diverse roles in mRNA metabolism. Progress in nucleic acid research and molecular biology. 2003;73:221-50.
- Chen J, Rappsilber J, Chiang Y-C, Russell P, Mann M, Denis CL. Purification and characterization of the 1.0 MDa CCR4-NOT complex identifies two novel components of the complex. Journal of molecular biology. 2001;314(4):683-94.
- Kruk JA, Dutta A, Fu J, Gilmour DS, Reese JC. The multifunctional Ccr4–Not complex directly promotes transcription elongation. Genes & development. 2011;25(6):581-93.
- Collart MA, Panasenko OO. The Ccr4–not complex. Gene. 2012;492(1):42-53.
- Chapat C, Corbo L. Novel roles of the CCR4–NOT complex. Wiley Interdisciplinary Reviews: RNA. 2014;5(6):883-901.
- Collart MA. Global control of gene expression in yeast by the Ccr4-Not complex. Gene. 2003;313:1-16.
- Collart MA. The Ccr4‐Not complex is a key regulator of eukaryotic gene expression. Wiley Interdisciplinary Reviews: RNA. 2016. 7(4):438-54
- Stowell JA, Webster MW, Kögel A, Wolf J, Shelley KL, Passmore LA. Reconstitution of targeted deadenylation by the Ccr4-Not complex and the YTH domain protein Mmi1. Cell reports. 2016;17(8):1978-89.
- Bhaskar V, Roudko V, Basquin J, Sharma K, Urlaub H, Séraphin B, Conti E. Structure and RNA-binding properties of the Not1–Not2–Not5 module of the yeast Ccr4–Not complex. Nature structural & molecular biology. 2013;20(11):1281-8.
- Buschauer R, Matsuo Y, Sugiyama T, Chen Y-H, Alhusaini N, Sweet T, Ikeuchi K, Cheng J, Matsuki Y, Nobuta R, et al. The Ccr4-Not complex monitors the translating ribosome for codon optimality. Science. 2020;368(6488).
- Collart MA, Struhl K. NOT1 (CDC39), NOT2 (CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes & development. 1994;8(5):525-37.
- Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104(3):377-86.
- Tucker M, Staples RR, Valencia‐Sanchez MA, Muhlrad D, Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. The EMBO journal. 2002;21(6):1427-36.
- Temme C, Zhang L, Kremmer E, Ihling C, Chartier A, Sinz A, Simonelig M, Wahle E. Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation. Rna. 2010;16(7):1356-70.
- Petit A-P, Wohlbold L, Bawankar P, Huntzinger E, Schmidt S, Izaurralde E, Weichenrieder O. The structural basis for the interaction between the CAF1 nuclease and the NOT1 scaffold of the human CCR4–NOT deadenylase complex. Nucleic acids research. 2012;40(21):11058-72.
- Basquin J, Roudko VV, Rode M, Basquin C, Séraphin B, Conti E. Architecture of the nuclease module of the yeast Ccr4-not complex: the Not1-Caf1-Ccr4 interaction. Molecular cell. 2012;48(2):207-18.
- Collart MA. The NOT4 RING E3 ligase: a relevant player in cotranslational quality control. ISRN molecular biology. 2013; 548359.
- Lau N, Kolkman A, van Schaik F, Mulder K, Pijnappel W, Heck A, Timmers H. Human Ccr4-Not complexes contain variable deadenylase subunits. Biochem J. 2009;422:443-53.
- Mostafa D, Takahashi A, Yanagiya A, Yamaguchi T, Abe T, Kureha T, Kuba K, Kanegae Y, Furuta Y, Yamamoto T, et al. Essential functions of the CNOT7/8 catalytic subunits of the CCR4-NOT complex in mRNA regulation and cell viability. RNA biology. 2020:1-14.
- Mauxion F, Prève B, Séraphin B. C2ORF29/CNOT11 and CNOT10 form a new module of the CCR4-NOT complex. RNA biology. 2013;10(2):267-76.
- Azzouz N, Panasenko OO, Deluen C, Hsieh J, Theiler G, Collart MA. Specific roles for the Ccr4-Not complex subunits in expression of the genome. Rna. 2009;15(3):377-83.
- Nasertorabi F, Batisse C, Diepholz M, Suck D, Böttcher B. Insights into the structure of the CCR4-NOT complex by electron microscopy. FEBS letters. 2011;585(14):2182-6.
- Boland A, Chen Y, Raisch T, Jonas S, Kuzuoğlu-Öztürk D, Wohlbold L, Weichenrieder O, Izaurralde E. Structure and assembly of the NOT module of the human CCR4–NOT complex. Nature structural & molecular biology. 2013;20(11):1289-97.
- Ukleja M, Cuellar J, Siwaszek A, Kasprzak JM, Czarnocki-Cieciura M, Bujnicki JM, Dziembowski A, Valpuesta JM. The architecture of the Schizosaccharomyces pombe CCR4-NOT complex. Nature communications. 2016;7:10433.
- Raisch T, Chang C-T, Levdansky Y, Muthukumar S, Raunser S, Valkov E. Reconstitution of recombinant human CCR4-NOT reveals molecular insights into regulated deadenylation. Nature communications. 2019;10(1):1-14.
- Raisch T, Sandmeir F, Weichenrieder O, Valkov E, Izaurralde E. Structural and biochemical analysis of a NOT1 MIF4G-like domain of the CCR4-NOT complex. Journal of Structural Biology. 2018;204(3):388-95.
- Collart MA, Panasenko OO. The Ccr4-Not complex: Architecture and structural insights. Macromolecular Protein Complexes: Springer; 2017. p. 349-79.
- Bawankar P, Loh B, Wohlbold L, Schmidt S, Izaurralde E. NOT10 and C2orf29/NOT11 form a conserved module of the CCR4-NOT complex that docks onto the NOT1 N-terminal domain. RNA biology. 2013;10(2):228-44.
- Albert TK, Lemaire M, van Berkum NL, Gentz R, Collart MA, Timmers HTM. Isolation and characterization of human orthologs of yeast CCR4–NOT complex subunits. Nucleic acids research. 2000;28(3):809-17.
- Hagkarim NC, Ryan EL, Byrd PJ, Hollingworth R, Shimwell NJ, Agathanggelou A, Vavasseur M, Kolbe V, Speiseder T, Dobner T, et al. Degradation of a novel DNA damage response protein, tankyrase 1 binding protein 1, following adenovirus infection. Journal of virology. 2018;92(12). e02034-17.
- Keskeny C, Raisch T, Sgromo A, Igreja C, Bhandari D, Weichenrieder O, Izaurralde E. A conserved CAF40-binding motif in metazoan NOT4 mediates association with the CCR4–NOT complex. Genes & development. 2019;33(3-4):236-52.
- Garces RG, Gillon W, Pai EF. Atomic model of human Rcd‐1 reveals an armadillo‐like‐repeat protein with in vitro nucleic acid binding properties. Protein science. 2007;16(2):176-88.
- Xu K, Bai Y, Zhang A, Zhang Q, Bartlam MG. Insights into the structure and architecture of the CCR4–NOT complex. Frontiers in genetics. 2014;5:137.
- Chen Y, Boland A, Kuzuoğlu-Öztürk D, Bawankar P, Loh B, Chang C-T, Weichenrieder O, Izaurralde E. A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Molecular cell. 2014;54(5):737-50.
- Ito K, Takahashi A, Morita M, Suzuki T, Yamamoto T. The role of the CNOT1 subunit of the CCR4-NOT complex in mRNA deadenylation and cell viability. Protein & cell. 2011;2(9):755-63.
- Shirai Y-T, Suzuki T, Morita M, Takahashi A, Yamamoto T. Multifunctional roles of the mammalian CCR4–NOT complex in physiological phenomena. Frontiers in genetics. 2014;5:286.
- Winkler GS. The mammalian anti‐proliferative BTG/Tob protein family. Journal of cellular physiology. 2010;222(1):66-72.
- Mauxion F, Faux C, Séraphin B. The BTG2 protein is a general activator of mRNA deadenylation. The EMBO journal. 2008;27(7):1039-48.
- Yang X, Morita M, Wang H, Suzuki T, Yang W, Luo Y, Zhao C, Yu Y, Bartlam M, Yamamoto T, et al. Crystal structures of human BTG2 and mouse TIS21 involved in suppression of CAF1 deadenylase activity. Nucleic acids research. 2008;36(21):6872-81.
- Miyasaka T, Morita M, Ito K, Suzuki T, Fukuda H, Takeda S, Inoue JI, Semba K, Yamamoto T. Interaction of antiproliferative protein Tob with the CCR4‐NOT deadenylase complex. Cancer science. 2008;99(4):755-61.
- Webster MW, Chen Y-H, Stowell JA, Alhusaini N, Sweet T, Graveley BR, Coller J, Passmore LA. mRNA deadenylation is coupled to translation rates by the differential activities of Ccr4-Not nucleases. Molecular cell. 2018;70(6):1089-100. e8.
- Braun JE, Huntzinger E, Fauser M, Izaurralde E. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Molecular cell. 2011;44(1):120-33.
- Chekulaeva M, Mathys H, Zipprich JT, Attig J, Colic M, Parker R, Filipowicz W. miRNA repression involves GW182-mediated recruitment of CCR4–NOT through conserved W-containing motifs. Nature structural & molecular biology. 2011;18(11):1218-26.
- Fabian MR, Cieplak MK, Frank F, Morita M, Green J, Srikumar T, Nagar B, Yamamoto T, Raught B, Duchaine TF, et al. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4–NOT. Nature structural & molecular biology. 2011;18(11):1211-1217.
- Mathys H, Basquin J, Ozgur S, Czarnocki-Cieciura M, Bonneau F, Aartse A, Dziembowski A, Nowotny M, Conti E, Filipowicz W. Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Molecular cell. 2014;54(5):751-65.
- Rouya C, Siddiqui N, Morita M, Duchaine TF, Fabian MR, Sonenberg N. Human DDX6 effects miRNA-mediated gene silencing via direct binding to CNOT1. Rna. 2014;20(9):1398-409.
- Ozgur S, Basquin J, Kamenska A, Filipowicz W, Standart N, Conti E. Structure of a human 4E-T/DDX6/CNOT1 complex reveals the different interplay of DDX6-binding proteins with the CCR4-NOT complex. Cell reports. 2015;13(4):703-11.
- Sgromo A, Raisch T, Backhaus C, Keskeny C, Alva V, Weichenrieder O, Izaurralde E. Drosophila Bag-of-marbles directly interacts with the CAF40 subunit of the CCR4–NOT complex to elicit repression of mRNA targets. Rna. 2018;24(3):381-95.
- Rodriguez-Gil A, Ritter O, Hornung J, Stekman H, Krüger M, Braun T, Kremmer E, Kracht M, Schmitz ML. HIPK family kinases bind and regulate the function of the CCR4-NOT complex. Molecular biology of the cell. 2016;27(12):1969-80.
- Yang C-Y, Ramamoorthy S, Boller S, Rosenbaum M, Gil AR, Mittler G, Imai Y, Kuba K, Grosschedl R. Interaction of CCR4–NOT with EBF1 regulates gene-specific transcription and mRNA stability in B lymphopoiesis. Genes & development. 2016;30(20):2310-24.
- Deluen C, James N, Maillet L, Molinete M, Theiler G, Lemaire M, Paquet N, Collart MA. The Ccr4-Not complex and yTAF1 (yTafII130p/yTafII145p) show physical and functional interactions. Molecular and cellular biology. 2002;22(19):6735-49.
This entry is adapted from the peer-reviewed paper 10.3390/cells9112379