Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
The importance of the circadian clock in maintaining human health is now widely acknowledged. Dysregulated and dampened clocks may be a common cause of age-related diseases and metabolic syndrome Thus, circadian clocks should be considered as therapeutic targets to mitigate disease symptoms.
- polymethoxy flavone
- ageing
- non-communicable diseases
- chemoprevention
1. Introduction—Circadian Clock
The 24 h rotation of the earth along its axis exposes all terrestrial organisms to light and dark cycles as well as to daily temperature fluctuations. In this environment, having a mechanism that can anticipate these changes instead of merely responding, provides an evolutionary advantage. Hence, an endogenous ≈ 24 h circadian clock (from the Latin circa diem meaning ‘about a day’) allows an organism to coordinate physiological activities according to cycling changes in the environment, food availability and predator risk [1]. Although initially observed in plants by Jean Jacques d’Ortous de Mairan (1729), the first genetic proof of circadian clock existence was made in 1971 by Konopka and Benzer who isolated the first arrhythmic Drosophila melanogaster mutants [2]. Fifteen years later, the period gene (per) was cloned in fruit flies [3] and another ten years later, the second circadian gene timeless (tim) was identified in fruit flies and soon thereafter also in mice [4][5]. Hall, Rosbash and Young, who were awarded the Nobel prize for physiology and medicine in 2017, elegantly demonstrated that the circadian rhythmicity is generated and sustained by transcriptional and translational feedback loops in which PER/TIM complexes inhibits their own CLOCK:CYCLE (CLK/CYC) activators [6][7][8]. A second interconnected feedback loop made by vrille and Pdp1ε (Par domain protein 1ε) sustains rhythmic transcription of clk, thus enhancing the stability of both cycles [9]. This model, initially described in fruit flies, was shown to be highly conserved across the kingdoms ranging from cyanobacteria and plants to insects and humans, although the function of some genes may have diverged between organisms [10]. In mammals, the CLOCK:BMAL1 heterodimer activates Per and Cryptochrome (Cry) transcription. Then, as in Drosophila, the PER:CRY complex translocates back to the nucleus to repress its transcription via CLOCK:BMAL1 interaction [11]. This primary negative feedback look is sustained by a second regulatory loop in which BMAL1 cyclic expression is maintained by the ROR (α, β and γ) activator and REV-ERB (α and β) repressor proteins [12] (Figure 1). Approximately 24 h are required to complete a full circadian cycle but several posttranscriptional and posttranslational events finely regulate these oscillators [13]. For example, casein kinase 1ε/δ (CK1ε/δ), plays a fundamental role in the establishment of a new circadian cycle by phosphorylating PER and CRY. Upon PER and CRY degradation by the 26 S proteasome complex, the suppression of CLOCK:BMAL1 activity is released allowing the cycle to start again [14]. Large gene expression profiling analyses have allowed the identification of several circadian-output genes [15][16]. While roughly 50% of genes in mammals show a level of circadian expression, researchers have highlighted that their profiles exhibit tissue-specific rhythms [17]. Strikingly, clocks have shown to regulate a multitude of pathways within the cells including their epigenetic profile, phosphorylation and metabolic profiles, and the microbiome in the organism [18][19][20][21][22].
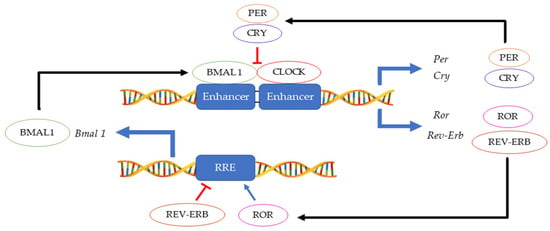
Figure 1. Main feedback loops regulating the circadian clock.
In mammals, as in other multicellular organisms, virtually all tissues possess circadian oscillators, making the system organisation highly complex. The light-entrainable pacemaker is in the suprachiasmatic nucleus (SCN) of the hypothalamus and its function is to synchronize peripherical clocks. While all these oscillate within a period close to 24 h, it is essential that they are synchronized with the external environmental conditions. Hence, the key function of the SCN clock is to receive environmental light information by retinohypothalamic track and synchronize other molecular oscillators, both within the SCN and in peripheral organs [23]. While the synchronization signal is transmitted by neurotransmitters and neuropeptides within the brain regions, hormonal secretion and neural innervation are used to synchronize the peripheral tissues [24]. Melatonin and glucocorticoids (Figure 2) are two examples of the manifestation of the circadian clock in mammals. Light information received from the SCN is transmitted to the pineal gland for nocturnal melatonin secretion. Circulating melatonin can entrain peripherical clocks interacting with molecular clock mechanisms acting as a signal for the dark phase of the photoperiod [25]. In the adrenal glands, the secretion of the hormone glucocorticoid—regulating glucose homeostasis—is also under circadian regulation. Indeed, cortisol concentration levels peak during the morning and during active periods in diurnal organisms, while its concentration is reduced during the sleeping phase [26]. While photic cues are the main circadian synchronisers for the SCN pacemaker, other non-photic zeitgebers such as arousal stimuli (e.g., social interaction or exercise), food/feeding regimes and temperature can also act as cues in the peripheral clocks. Moreover, in this complex network, a hormone and metabolic signal-based bi-directional communication exists between the SCN and non-SCN oscillators, providing plasticity to the system and optimal adaptation to the environment [27][28].
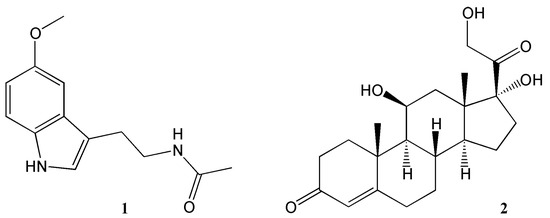
Figure 2. Key hormones regulating circadian rhythms. 1 = melatonin, 2 = corticosterone.
Due to the impact of circadian oscillators on the physiology and behaviour of organisms, their dysregulations and disruptions are associated with the development of diverse pathologies. In mouse models, mutations or deletions in core circadian genes (e.g., BMAL1) cause increased levels of glucose and lipids, leading to premature ageing [29]. Interestingly, BMAL1 downregulation leads to tissue-specific dysfunctions causing metabolic and triglyceride biosynthesis impairments in the muscle, or obesity when knocked-down in adipose tissue [30][31]. Similarly, surgical removal of the SCN is linked to the onset of tumour growth and alteration in the microbiota and immune cells, in the intestine of mouse models [32][33]. In humans, one of the major contributions to circadian disorders is the misalignment between the endogenous clock and the environmental rhythms (such as the day-night cycle). Artificial light, shift work, travel and social lags are all clock misalignments introduced by the modern lifestyle. For example, night shift workers show an increased risk to develop several types of cancer, cardiovascular and metabolic disorders, psychiatric disorders, obesity and type-2 diabetes [34][35][36]. Traveling across multiple time zones, causes circadian rhythm desynchronization which ultimately leads to changes in sleep architecture, mood, hormone profiles, and gastrointestinal dysfunctions [37]. A great proportion of the teenage population experience social jet lag in which a variation in sleep pattern is observed between school/work days versus school/work free days. Notably, social jet lag is associated with an elevated consumption of alcohol and tobacco, as well as a higher incidence of obesity, diabetes, cancer, and cardiovascular disease [38][39]. Because circadian clocks regulate several cellular mechanisms such as oxidative stress, inflammation, neurotransmitter biosynthesis and metabolism, they have been linked to the development and progression of human neurodegenerative disorders [40]. While at present, the bi-directional relation between circadian disruption and neurodegeneration is not fully understood, evidence indicates that desynchronization of the clock over a lifetime, enhances the deposition of misfolded protein aggregates in Alzheimer’s and Parkinson’s diseases [41]. Moreover, in humans, single-nucleotide polymorphisms in core clock genes (e.g., CLOCK, BMAL1 and PER1) have been shown to increase the risk of Alzheimer’s and Parkinson’s Disease development [42].
2. Effects of Natural Products on Circadian Rhythm
Phytochemicals, compounds derived from plants, have been known to influence a wide range of pharmacological processes. Many phytochemical compounds, such as flavonoids, alkaloids, polyphenols and melatonin, have been reported to have a regulatory effect on expression of genes linked to the circadian clock, and are thus expected to play a role in regulating the internal environment [43]. Flavonoids (Figure 3) in particular raised interest as compounds that may affect circadian rhythm and diseases related to appropriate regulation of circadian rhythm [44]. In spite of a wealth of information showcasing the role of flavonoids in various disease states, very little work has been done so far to specifically ascertain the effects of these molecules on the circadian physiology. Nevertheless, these studies are a rich source of information, and help us understand the significant role played by flavonoids in modulating different circadian systems. A flavonoid-rich fraction of the plant Cyclocarya paliurus was shown to have modulatory effects on both the liver clock genes as well as intestinal microbiota in a circadian rhythm disorder mice model. A robust rhythmic expression in most liver clock genes was observed, mainly Clock1, Per1, Per2, Per3, Bmal1, Sirt1, Cry1 and Cry2 over a 24 h period. Analysis of the plant extract showed kaempferol-3-O-β-glucuronide as the predominant flavonoid, with kaempferol-3-O-α-l-rhamnopyranoside, quercetin and quercetin-3-O-glucoside (isoquercitrin) found in varying proportions [45]. Other studies investigating the effect of flavonoids on circadian rhythms in different mice models showed similar outcomes [46][47][48][49]. Most flavonoids have poor bioavailability in mammals, and merely pass through the digestive tract to be metabolised by intestinal microbes in the colon. Therefore, some authors leave open the option that flavonoids may not directly affect mammalian physiology but do so indirectly by altering the gut microbiota which then, in turn, produce metabolites that affect the mammalian circadian system [45][49]. The concept of the gut-brain axis is intriguing and attracting increasing interest, though much still remains to be further explored.
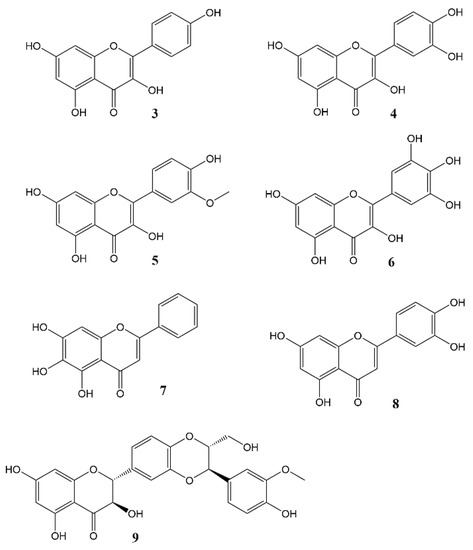
Figure 3. Flavonols and flavones 3 = Kaempferol, 4 = Quercetin, 5 = Isorhamnetin, 6 = Myricetin, 7 = Baicalein, 8 = Luteolin, 9 = Silybin A.
Interestingly, mutations that alter Arabidopsis flavonoid metabolism, concomitantly affect the plant’s circadian clock [50]. This indicates that flavonoids can affect circadian rhythms in both the animal and plant kingdom, which may be seen as further confirmation that the mechanism of the circadian clock is highly conserved. Some flavonoids and their effects on the circadian variations are expanded on below:
The bioflavonoid quercetin has a significant effect on the sleep-wake cycle in male Sprague Dawley rats [51]. When sleep-wake states were classified as wakefulness, rapid eye movement (REM) sleep and non-REM sleep, a post hoc analysis of the rat’s diurnal cycle after quercetin administration (200 mg/kg) showed a notable decrease in REM sleep during the first three hours, an effect which was seen almost immediately. Additionally, a decrease in wakefulness and increased non-REM sleep during the last four hours were observed. The proposed pathway through which quercetin exerts its effects was the GABAergic pathway and GABA-independent mechanisms. Similarly, baicalein, a flavonoid found in Scutellaria baicalensis, affects the same pathway. It was shown to promote increase in sleep time via GABAergic non-benzodiazepine sites in mouse brains [52][53][54].
3. Effects of Nobiletin Circadian Rhythm and Metabolism
Nobiletin (Figure 4) is a polymethoxy flavone that almost uniquely accumulates in the peel of Citrus fruits. Dried citrus peel, known as Citri Reticulatae Pericarpium or Chenpi in Chinese has traditionally been used to promote the circulation of qi (energy) throughout the body. The concept of ‘qi’ does not translate easily into western concepts of pharmacology, but Chenpi is used to sooth emotions including anger and irritability, and to supplement treatment of indigestion, and abdominal fullness through promotion of gastrointestinal motility [55].
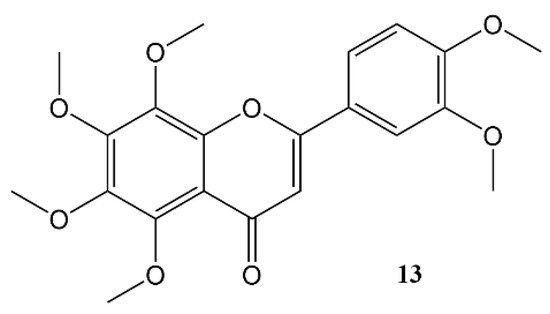
Figure 4. Nobiletin.
A considerable body of work, done at the Department of Biochemistry and Molecular Biology of the University of Texas Health Science Center at Houston, has indicated that nobiletin can have a beneficial effect on human metabolism and acts through modulation of circadian clocks. Using a high-throughput chemical screening assay, based on fibroblasts expressing a PER2::Luc reporter gene construct [56][57], nobiletin was identified as a particularly effective clock amplitude-enhancing small molecule. Moreover, the assay shows that this flavone can directly affect the mammalian circadian system [43][58]. Furthermore, pharmacokinetic studies revealed significant brain and systemic exposure to nobiletin is feasible after oral administration [43]. We might speculate that this lipohilic flavone with its planar structure may mimick the activity of corticosterone (Figure 1), much like flavonoid phytoestrogens are known to mimick the activity of gonadocorticoids.
This entry is adapted from the peer-reviewed paper 10.3390/molecules27227727
References
- Dunlap, J.C. Molecular Bases for Circadian Clocks. Cell 1999, 96, 271–290.
- Konopka, R.J.; Benzer, S. Clock Mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 1971, 68, 2112–2116.
- Reddy, P.; Zehring, W.A.; Wheeler, D.A.; Pirrotta, V.; Hadfield, C.; Hall, J.C.; Rosbash, M. Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell 1984, 38, 701–710.
- Koike, N.; Hida, A.; Numano, R.; Hirose, M.; Sakaki, Y.; Tei, H. Identification of the mammalian homologues of the Drosophila timeless gene, Timeless1. FEBS Lett. 1998, 441, 427–431.
- Sehgal, A.; Price, J.L.; Man, B.; Young, M.W. Loss of Circadian Behavioral Rhythms and per RNA Oscillations in the Drosophila Mutant timeless. Science 1994, 263, 1603–1606.
- Hardin, P.E.; Hall, J.C.; Rosbash, M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 1990, 343, 536–540.
- Lee, C.; Bae, K.; Edery, I. PER and TIM Inhibit the DNA Binding Activity of a Drosophila CLOCK-CYC/dBMAL1 Heterodimer without Disrupting Formation of the Heterodimer: A Basis for Circadian Transcription. Mol. Cell. Biol. 1999, 19, 5316–5325.
- Sehgal, A.; Rothenfluh-Hilfiker, A.; Hunter-Ensor, M.; Chen, Y.; Myers, M.P.; Young, M.W. Rhythmic Expression of timeless: A Basis for Promoting Circadian Cycles in period Gene Autoregulation. Science 1995, 270, 808–810.
- Cyran, S.A.; Buchsbaum, A.M.; Reddy, K.L.; Lin, M.-C.; Glossop, N.R.; Hardin, P.E.; Young, M.W.; Storti, R.V.; Blau, J. vrille, Pdp1, and dClock Form a Second Feedback Loop in the Drosophila Circadian Clock. Cell 2003, 112, 329–341.
- Bhadra, U.; Thakkar, N.; Das, P.; Bhadra, M.P. Evolution of circadian rhythms: From bacteria to human. Sleep Med. 2017, 35, 49–61.
- Sato, T.K.; Yamada, R.G.; Ukai, H.; Baggs, J.E.; Miraglia, L.J.; Kobayashi, T.J.; Welsh, D.K.; Kay, S.A.; Ueda, H.; Hogenesch, J.B. Feedback repression is required for mammalian circadian clock function. Nat. Genet. 2006, 38, 312–319.
- Takahashi, J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017, 18, 164–179.
- Dubowy, C.; Sehgal, A. Circadian Rhythms and Sleep in Drosophila melanogaster. Genetics 2017, 205, 1373–1397.
- Akashi, M.; Tsuchiya, Y.; Yoshino, T.; Nishida, E. Control of Intracellular Dynamics of Mammalian Period Proteins by Casein Kinase I ε (CKIε) and CKIδ in Cultured Cells. Mol. Cell. Biol. 2002, 22, 1693–1703.
- Ruben, M.D.; Wu, G.; Smith, D.F.; Schmidt, R.E.; Francey, L.J.; Lee, Y.Y.; Anafi, R.C.; Hogenesch, J.B. A database of tissue-specific rhythmically expressed human genes has potential applications in circadian medicine. Sci. Transl. Med. 2018, 10, eaat8806.
- Zhang, R.; Lahens, N.F.; Ballance, H.I.; Hughes, M.E.; Hogenesch, J.B. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. USA 2014, 111, 16219–16224.
- Storch, K.-F.; Lipan, O.; Leykin, I.; Viswanathan, N.; Davis, F.C.; Wong, W.H.; Weitz, C.J. Extensive and divergent circadian gene expression in liver and heart. Nature 2002, 417, 78–83.
- Bishehsari, F.; Voigt, R.M.; Keshavarzian, A. Circadian rhythms and the gut microbiota: From the metabolic syndrome to cancer. Nat. Rev. Endocrinol. 2020, 16, 731–739.
- Chiang, C.-K.; Xu, B.; Mehta, N.; Mayne, J.; Sun, W.Y.L.; Cheng, K.; Ning, Z.; Dong, J.; Zou, H.; Cheng, H.-Y.M.; et al. Phosphoproteome Profiling Reveals Circadian Clock Regulation of Posttranslational Modifications in the Murine Hippocampus. Front. Neurol. 2017, 8, 110.
- Malik, D.M.; Paschos, G.K.; Sehgal, A.; Weljie, A.M. Circadian and Sleep Metabolomics Across Species. J. Mol. Biol. 2020, 432, 3578–3610.
- Robles, M.S.; Humphrey, S.J.; Mann, M. Phosphorylation Is a Central Mechanism for Circadian Control of Metabolism and Physiology. Cell Metab. 2017, 25, 118–127.
- Wang, J.; Mauvoisin, D.; Martin, E.; Atger, F.; Galindo, A.N.; Dayon, L.; Sizzano, F.; Palini, A.; Kussmann, M.; Waridel, P.; et al. Nuclear Proteomics Uncovers Diurnal Regulatory Landscapes in Mouse Liver. Cell Metab. 2017, 25, 102–117.
- Finger, A.-M.; Dibner, C.; Kramer, A. Coupled network of the circadian clocks: A driving force of rhythmic physiology. FEBS Lett. 2020, 594, 2734–2769.
- Buijs, R.M.; Ruiz, M.A.G.; Hernández, R.M.; Cortés, B.R. The suprachiasmatic nucleus; A responsive clock regulating homeostasis by daily changing the setpoints of physiological parameters. Auton. Neurosci. 2019, 218, 43–50.
- Hardeland, R.; Madrid, J.A.; Tan, D.-X.; Reiter, R.J. Melatonin, the circadian multioscillator system and health: The need for detailed analyses of peripheral melatonin signaling. J. Pineal Res. 2012, 52, 139–166.
- Ishida, A.; Mutoh, T.; Ueyama, T.; Bando, H.; Masubuchi, S.; Nakahara, D.; Tsujimoto, G.; Okamura, H. Light activates the adrenal gland: Timing of gene expression and glucocorticoid release. Cell Metab. 2005, 2, 297–307.
- Pilorz, V.; Astiz, M.; Heinen, K.O.; Rawashdeh, O.; Oster, H. The Concept of Coupling in the Mammalian Circadian Clock Network. J. Mol. Biol. 2020, 432, 3618–3638.
- Zhang, S.; Dai, M.; Wang, X.; Jiang, S.-H.; Hu, L.-P.; Zhang, X.-L.; Zhang, Z.-G. Signalling entrains the peripheral circadian clock. Cell. Signal. 2020, 69, 109433.
- Yang, G.; Chen, L.; Grant, G.R.; Paschos, G.; Song, W.-L.; Musiek, E.S.; Lee, V.; McLoughlin, S.C.; Grosser, T.; Cotsarelis, G.; et al. Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci. Transl. Med. 2016, 8, 324ra16.
- Dyar, K.A.; Hubert, M.J.; Mir, A.A.; Ciciliot, S.; Lutter, D.; Greulich, F.; Quagliarini, F.; Kleinert, M.; Fischer, K.; Eichmann, T.O.; et al. Transcriptional programming of lipid and amino acid metabolism by the skeletal muscle circadian clock. PLoS Biol. 2018, 16, e2005886.
- Paschos, G.K.; Ibrahim, S.; Song, W.-L.; Kunieda, T.; Grant, G.; Reyes, T.M.; Bradfield, C.A.; Vaughan, C.H.; Eiden, M.; Masoodi, M.; et al. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat. Med. 2012, 18, 1768–1777.
- Filipski, E.; King, V.M.; Li, X.; Granda, T.; Mormont, M.-C.; Liu, X.; Claustrat, B.; Hastings, M.H.; Lévi, F. Host Circadian Clock as a Control Point in Tumor Progression. JNCI J. Natl. Cancer Inst. 2002, 94, 690–697.
- Godinho-Silva, C.; Domingues, R.G.; Rendas, M.; Raposo, B.; Ribeiro, H.; da Silva, J.A.; Vieira, A.; Costa, R.M.; Barbosa-Morais, N.L.; Carvalho, T.; et al. Light-entrained and brain-tuned circadian circuits regulate ILC3s and gut homeostasis. Nature 2019, 574, 254–258.
- Bedrosian, T.A.; Nelson, R.J. Timing of light exposure affects mood and brain circuits. Transl. Psychiatry 2017, 7, e1017.
- Morris, C.J.; Purvis, T.E.; Mistretta, J.; Scheer, F.A.J.L. Effects of the Internal Circadian System and Circadian Misalignment on Glucose Tolerance in Chronic Shift Workers. J. Clin. Endocrinol. Metab. 2016, 101, 1066–1074.
- Sinturel, F.; Petrenko, V.; Dibner, C. Circadian Clocks Make Metabolism Run. J. Mol. Biol. 2020, 432, 3680–3699.
- Sack, R.L. Jet Lag. N. Engl. J. Med. 2010, 362, 440–447.
- Giuntella, O.; Mazzonna, F. Sunset time and the economic effects of social jetlag: Evidence from US time zone borders. J. Health Econ. 2019, 65, 210–226.
- Roenneberg, T.; Allebrandt, K.V.; Merrow, M.; Vetter, C. Social Jetlag and Obesity. Curr. Biol. 2012, 22, 939–943.
- Nassan, M.; Videnovic, A. Circadian rhythms in neurodegenerative disorders. Nat. Rev. Neurol. 2022, 18, 7–24.
- Kim, M.; Subramanian, M.; Cho, Y.-H.; Kim, G.-H.; Lee, E.; Park, J.-J. Short-term exposure to dim light at night disrupts rhythmic behaviors and causes neurodegeneration in fly models of tauopathy and Alzheimer’s disease. Biochem. Biophys. Res. Commun. 2018, 495, 1722–1729.
- Logan, R.W.; McClung, C.A. Rhythms of life: Circadian disruption and brain disorders across the lifespan. Nat. Rev. Neurosci. 2019, 20, 49–65.
- Liu, F.; Zhang, X.; Zhao, B.; Tan, X.; Wang, L.; Liu, X. Role of Food Phytochemicals in the Modulation of Circadian Clocks. J. Agric. Food Chem. 2019, 67, 8735–8739.
- Xu, T.; Lu, B. The effects of phytochemicals on circadian rhythm and related diseases. Crit. Rev. Food Sci. Nutr. 2019, 59, 882–892.
- Song, D.; Ho, C.-T.; Zhang, X.; Wu, Z.; Cao, J. Modulatory effect of Cyclocarya paliurus flavonoids on the intestinal microbiota and liver clock genes of circadian rhythm disorder mice model. Food Res. Int. 2020, 138, 109769.
- He, B.; Nohara, K.; Park, N.; Park, Y.-S.; Guillory, B.; Zhao, Z.; Garcia, J.M.; Koike, N.; Lee, C.C.; Takahashi, J.S.; et al. The Small Molecule Nobiletin Targets the Molecular Oscillator to Enhance Circadian Rhythms and Protect against Metabolic Syndrome. Cell Metab. 2016, 23, 610–621.
- Qi, G.; Mi, Y.; Liu, Z.; Fan, R.; Qiao, Q.; Sun, Y.; Ren, B.; Liu, X. Dietary tea polyphenols ameliorate metabolic syndrome and memory impairment via circadian clock related mechanisms. J. Funct. Foods 2017, 34, 168–180.
- Toda, K.; Hitoe, S.; Takeda, S.; Shimizu, N.; Shimoda, H. Passionflower Extract Induces High-amplitude Rhythms without Phase Shifts in the Expression of Several Circadian Clock Genes in Vitro and in Vivo. Int. J. Biomed. Sci. 2017, 13, 84–92.
- Zhao, K.; Ge, Q.; Zhang, X.; Shao, X.; Wei, Y.; Wang, H.; Xu, F. Genomic analysis of intestinal flora and liver genes in mice with circadian rhythm disorders fed with flavonoids from Sedum aizoon L. Food Biosci. 2022, 50, 102067.
- Hildreth, S.B.; Littleton, E.S.; Clark, L.C.; Puller, G.C.; Kojima, S.; Winkel, B.S.J. Mutations that alter Arabidopsis flavonoid metabolism affect the circadian clock. Plant J. 2022, 110, 932–945.
- Kambe, D.; Kotani, M.; Yoshimoto, M.; Kaku, S.; Chaki, S.; Honda, K. Effects of quercetin on the sleep–wake cycle in rats: Involvement of gamma-aminobutyric acid receptor type A in regulation of rapid eye movement sleep. Brain Res. 2010, 1330, 83–88.
- Chang, H.-H.; Yi, P.-L.; Cheng, C.-H.; Lu, C.-Y.; Hsiao, Y.-T.; Tsai, Y.-F.; Li, C.-L.; Chang, F.-C. Biphasic effects of baicalin, an active constituent of Scutellaria baicalensis Georgi, in the spontaneous sleep-wake regulation. J. Ethnopharmacol. 2011, 135, 359–368.
- de Carvalho, R.S.M.; Duarte, F.S.; de Lima, T.C.M. Involvement of GABAergic non-benzodiazepine sites in the anxiolytic-like and sedative effects of the flavonoid baicalein in mice. Behav. Brain Res. 2011, 221, 75–82.
- Hong, K.-B.; Han, S.H.; Park, Y.; Suh, H.J.; Choi, H.-S. Romaine Lettuce/Skullcap Mixture Improves Sleep Behavior in Vertebrate Models. Biol. Pharm. Bull. 2018, 41, 1269–1276.
- Yu, X.; Sun, S.; Guo, Y.; Liu, Y.; Yang, D.; Li, G.; Lü, S. Citri Reticulatae Pericarpium (Chenpi): Botany, ethnopharmacology, phytochemistry, and pharmacology of a frequently used traditional Chinese medicine. J. Ethnopharmacol. 2018, 220, 265–282.
- Chen, Z.; Yoo, S.-H.; Takahashi, J.S. Small molecule modifiers of circadian clocks. Cell. Mol. Life Sci. 2013, 70, 2985–2998.
- Chen, Z.; Yoo, S.-H.; Park, Y.-S.; Kim, K.-H.; Wei, S.; Buhr, E.; Ye, Z.-Y.; Pan, H.-L.; Takahashi, J.S. Identification of diverse modulators of central and peripheral circadian clocks by high-throughput chemical screening. Proc. Natl. Acad. Sci. USA 2012, 109, 101–106.
- Shinozaki, A.; Misawa, K.; Ikeda, Y.; Haraguchi, A.; Kamagata, M.; Tahara, Y.; Shibata, S. Potent Effects of Flavonoid Nobiletin on Amplitude, Period, and Phase of the Circadian Clock Rhythm in PER2::LUCIFERASE Mouse Embryonic Fibroblasts. PLoS ONE 2017, 12, e0170904.
This entry is offline, you can click here to edit this entry!
