Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
COVID-19 is no longer a global pandemic due to development and integration of different technologies for the diagnosis and treatment of the disease, technological advancement in the field of molecular biology, electronics, computer science, artificial intelligence, Internet of Things, nanotechnology, etc. has led to the development of molecular approaches and computer aided diagnosis for the detection of COVID-19.
- biosensors
- COVID-19
- artificial intelligence
1. Introduction
The year 2020 has witnessed a massive global burden due to the spread of the pneumonia causing virus known as SARS-CoV2 or COVID-19. The disease has led to massive screening, quarantines, restriction of movement, closure of land and borders, lockdowns, closure of educational, sportive and entertainment centers, and forced people to work from home [1][2]. In order to control the disease, scientists from different fields work hand in hand together to develop diagnostic approaches, prediction models, treatment control strategies, vaccines, etc. Screening of SARS-CoV-2 using a lab-bench assay is regarded as the first line of action in terms of minimizing spread and allowing for early treatment of the disease. This prompted the Chinese government to enact several testing points [2][3][4].
Medical experts rely on two main molecular approaches, which include RT-PCR and antibody–antigen based techniques, for the detection of the disease. However, among these two molecular testing approaches, RT-PCR is regarded as the gold standard technique due to it specificity and accuracy. The tests allow healthcare experts to detect viral nucleic acid from patient samples collected using a nasal swab, which is amplified using PCR machine. Antigen–antibody revolves around the binding between a synthesized recombinant antigen and the antibodies present in the body, which elicit an antigen–antibody reaction [5][6][7].
Even though molecular techniques such as RT-PCR and antibody testing are regarded as the standard procedures for the detection of SARS-CoV-2, they are hindered by several challenges, which include the incidence of false positive results, which can lead to misdiagnosis. These testing procedures are also costly, especially in remote areas and countries with substandard healthcare systems. As an alternative, healthcare professionals employ radiographic screening using X-ray imaging and CT-scan imaging, which allow scientists to discriminate between positive and negative cases. Others employ these techniques as a follow-up approach or as confirmation tests. As a result of massive or large-scale screening of radiographic images, these techniques can be tedious for radiologists and can led to misinterpretation [3][4][8][9].
In order to address these issues, scientists merge radiographic imaging with computer applications to develop computer-aided diagnosis (CAD), which allows screening of thousands of images with high accuracy, precision, and specificity [10][11]. CAD has been shown to aid medical experts in the past in the detection of different types of cancer, such as breast cancer [12], colon cancer [13], prostate cancer [14], brain cancer [15], tuberculosis [16], bacterial pneumonia [17], non-COVID-19 viral pneumonia [18], and skin diseases [19].
The integration of IoT with medical care, known as IoMT, is changing the landscape of patient care, diagnosis, and treatment. IoMT revolves around the interconnection between medical devices using internet. The platform enables machine–machine communication, patient–machine communication, machine–medical professional communication, etc. Examples of IoMT system include patient tracking devices, remote patient monitoring, medication tracking devices, etc. [20][21][22].
The prospect of smart diagnosis has been gaining ground in the last decade. The integration of smart technologies, such as AI and IoMT, with conventional diagnostic approaches has the potential to improve diagnosis and real-time, or point-of-care, detection, as well as to minimize errors and allow the sharing of medical data between devices, end users, and hospital cloud systems [23][24].
2. COVID-19
For the first time in a century, the world witnessed another global pandemic caused by a coronavirus known as Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-2). The virus is traced back to a sea food market in Wuhan, Hubei province, China, in late December, 2019. Coronaviruses are positive, single-stranded RNA viruses which belong to the Coronaviridae family, which also include SARS-CoV-1 and MERS-CoV. Both the two coronaviruses have caused epidemics and endemics in the last few years [1][25].
SARS-CoV-1 was first identified in the year 2003 in China, where bats are regarded as the main reservoirs. The virus has spread to four other countries, causing global epidemics. SARS-CoV-1 affected close to 8000 people with an approximately 10% mortality rate. The World Health Organization, along with other international and non-Governmental organizations, collaborated to control and prevent further spread of the virus [26][27].
Middle East Respiratory Syndrome Coronavirus (MERS-CoV) is another virus that belongs to Coronaviridae family that caused a global burden in the year 2012. The virus was first identified in Saudi Arabia, and later spread to 27 countries, leading to approximately 2600 cases. The disease has infected thousands, and approximately 35% of patients died from the disease. Dromedary camels were linked with the transmission of the virus to human primates [28][29]. Figure 1 shows the differences and similarities between SARS-CoV-1, MERS-CoV-2, and SERS-CoV-2 in terms of number of cases, fatality rate, mortality rate, reservoir, etc.
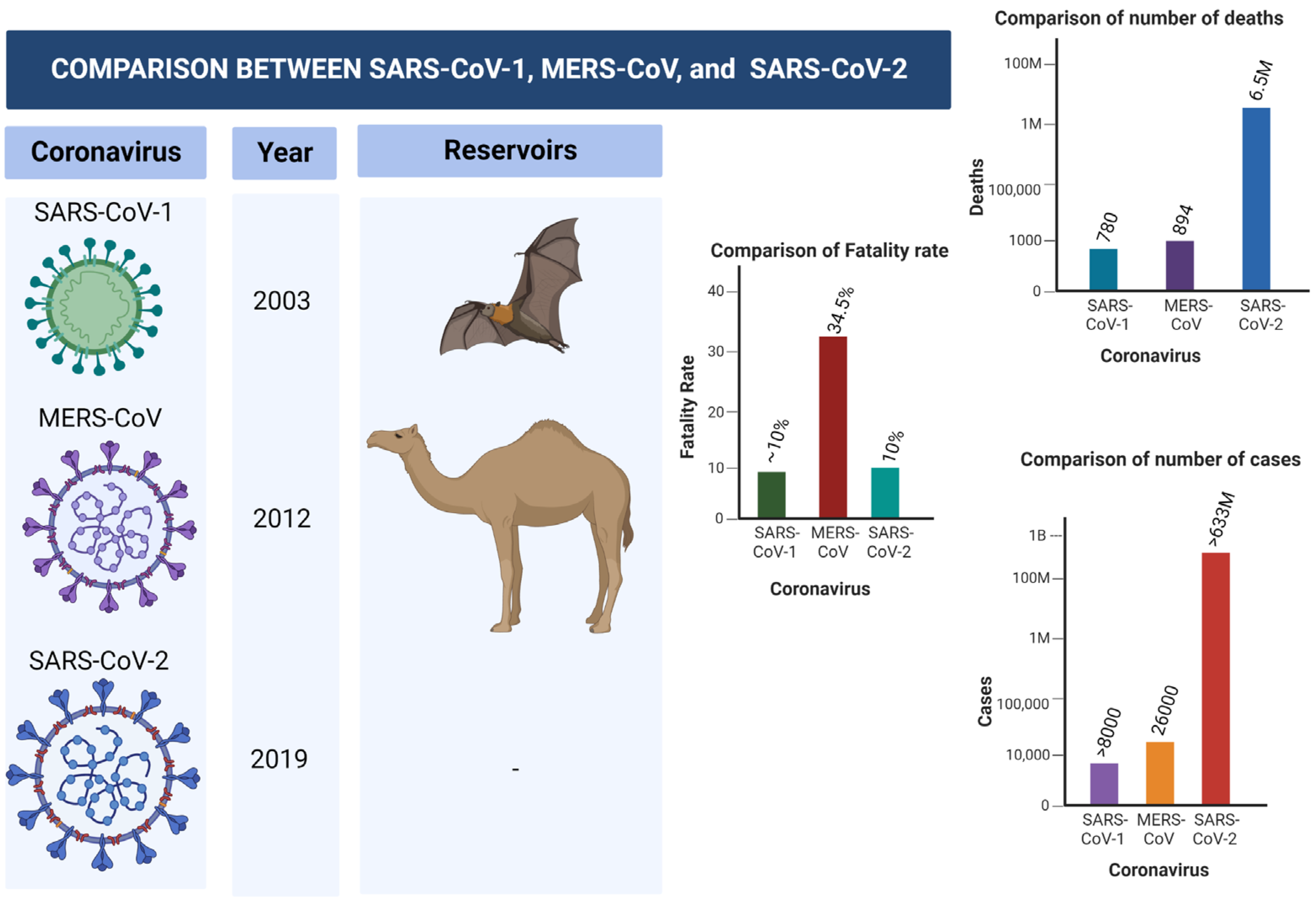
Figure 1. Comparison between SARS-CoV-1, MERS-CoV-2, and SARS-CoV-2.
2.1. Transmission of SARS-CoV-2
Unlike previous coronavirus diseases that are associated with animal transmission, such as bat in SARS-CoV-1 and dromedary camels in MERS-CoV, no animal reservoir has been found for SARS-CoV-2 [30]. Several studies have shown that the virus can be spread directly from one person to another (Human–human transmission) via sneezing or coughing, or indirectly, such as by coming in contact with surfaces infected with the virus [30][31].
2.2. Symptoms of SARS-CoV-2
The clinical spectrum of the COVID-19 disease ranges from asymptomatic to severe acute respiratory disease and death. People infected with the disease display pneumonia symptoms, which include shortness of breath, sore throat, fever, fatigue, and cough. People that are at risk of COVID-19 include elderly people who are suffering from chronic diseases, such as chronic lung disease, cancer, hypertension, renal and kidney diseases, diabetes, cardiovascular diseases, etc. [31][32]. The clinical manifestations of SARS-CoV-2 are presented in Figure 2 (redesigned using Biorender [33]).
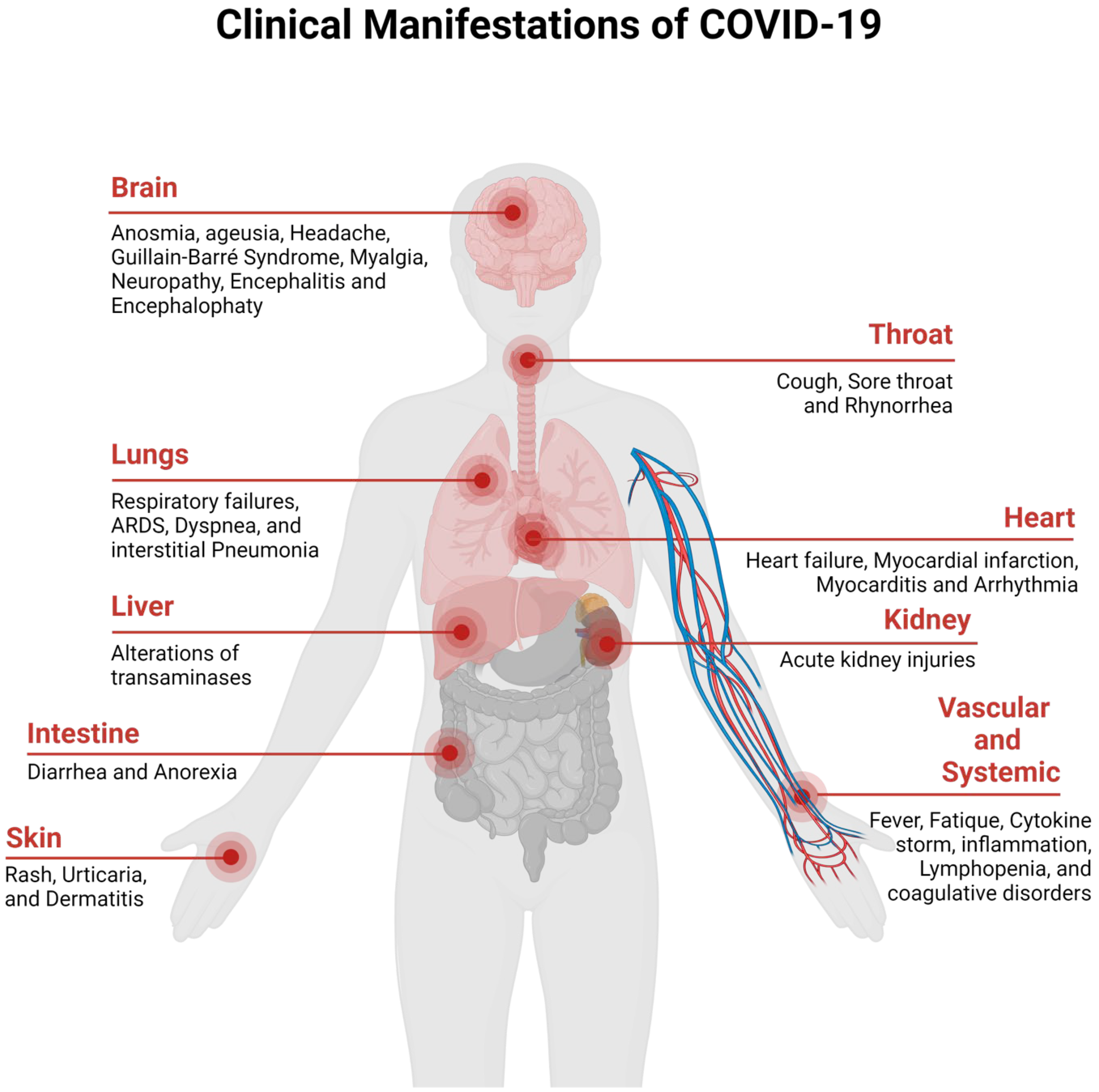
Figure 2. Clinical Manifestation of COVID-19.
3. Molecular Diagnosis of COVID-19
Early and accurate detection of COVID-19 disease is crucial for timely management and prevention. The field of disease detection has been transformed from conventional diagnosis, such as microscopy, with lower sensitivity and specificity, to molecular diagnosis, such as antigen–antibody, enzyme-substrate and NA probe-target, and biosensing technologies [5][34], as shown in Figure 3. Diagnostic imaging based on CT scans, X-rays and ultrasound imaging are currently in use as an alternative or confirmatory approach for the detection of COVID-19 [35][36].
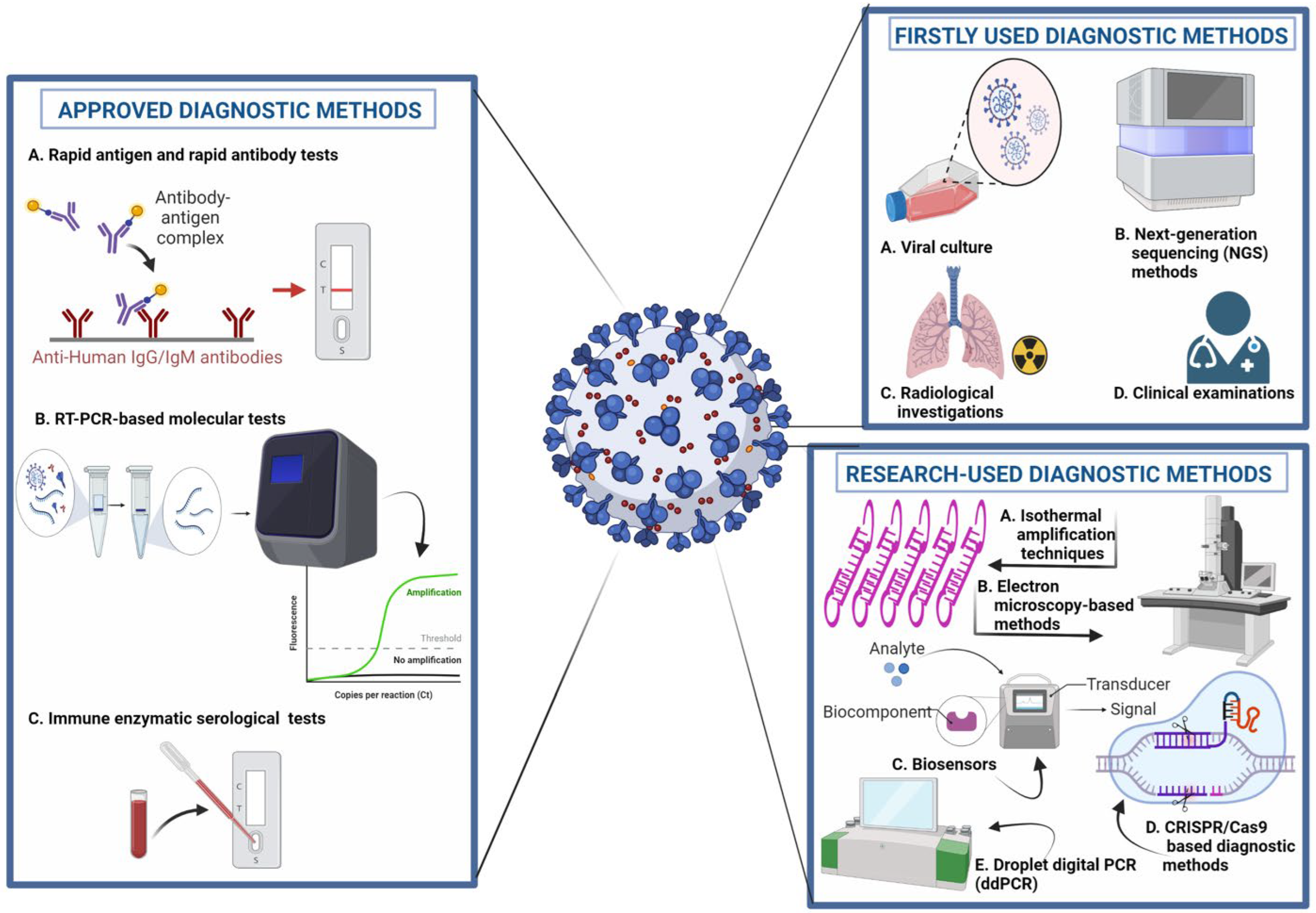
Figure 3. Molecular diagnostic approaches for the detection of COVID-19. 1. Approved Diagnostic Methods; A. Rapid Antigen and Rapid Antibody Tests; B. RT-PCR-based Molecular Tests; C. Immune Enzymatic Serological Tests. 2. Firstly Used Diagnostic; A. Viral Culture; B. Next Generation Sequencing (NGS) Methods; C. Radiological Investigation; D. Clinical Examination; 3. Research-used Diagnostic Methods; A. Isothermal Amplification Techniques; B. Electron microscopy-based Methods; C. Biosensors; D. CRISPR/Cas9-based Diagnostic Methods. E. Droplet digital PCR (ddPCR).
3.1. Laboratory Assays
Accurate, sensitive, and rapid laboratory assays for the detection of SARS-CoV-2 are crucial for the treatment and control of COVID-19 infection. Currently, there are a myriad of tests available in the market. However, the adoption of appropriate laboratory testing techniques and types of specimen (nasopharyngeal aspirates, nasopharyngeal swabs, mid-turbinate swabs, oropharyngeal swab, etc.) is one of the cornerstones for the timely management and control of the disease. The literature encompasses several studies focusing on laboratory testing procedures, which include nucleic acid amplifications, antigen tests, antibody tests, and point-of-care testing. These procedures are currently employed in the detection of SARS-CoV-2 in clinical diagnosis of symptomatic patients, asymptomatic population screening, contact investigations, targeted high-risk population screening, retrospective population screening, monitoring of infectivity, disease severity monitoring, etc. [35][36][37].
3.1.1. Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
RT-PCR is regarded as the most reliable approach for the detection SARS-CoV-2 [5][38][39]. RT-PCR is a nuclear-derived approach for the detection of the genetic content of pathogens, such as viruses (such as Zika and Ebola) and bacteria. The early RT-PCR testing employ radioisotope markers which are subsequently replaced by fluorescent dyes. As shown in Figure 4, the procedure for conducting RT-PCR test follows four steps:
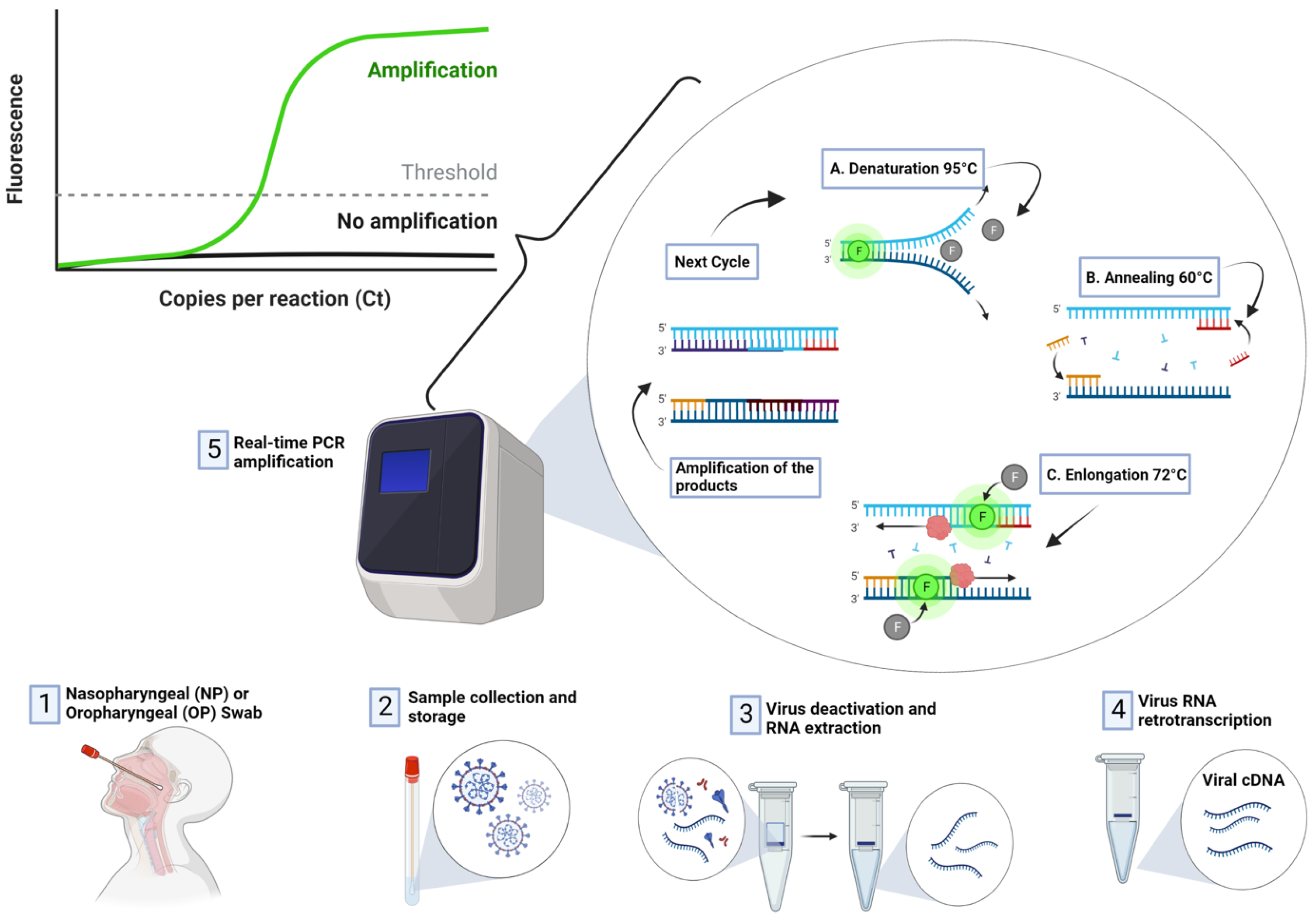
Figure 4. Detection of COVID-19 using RT-PCR Technique.
Sample collection: Nasal swab and nasopharyngeal samples are collected by medical experts. The samples are sealed and transported to the laboratory for detection.
Extraction: This step allows medical technologies to extract or isolate viral NA. This stage revolves around the use of chemicals to remove components such as fats and proteins.
PCR: The isolated viral NA is further amplified using a PCR machine, also known as a thermal cycler. PCR machines amplify thousands of complies of the viral NA, which increases the sensitivity and specificity of detection. Reverse transcription is carried out in order to convert RNA strands of the virus to DNA.
Detection: After the RNA is transcribed to DNA and amplified, the machine detects the presence of virus DNA due to the release of fluorescent dye, which can be measured in real-time and presented on the computer screen.
The RT-PCR method is highly specific and sensitive compared to the antigen–antibody method. The test can take between 3 and 6 h to process and obtain results. Moreover, this approach has shown to be faster, reliable, and to present a lower rate of errors or false positive results compared to other approaches. One of the disadvantages of RT-PCR is that it can’t be used to identify past diseases, which is crucial for understanding the pathology and spread of the diseases [40][41].
3.1.2. Antibody-Based Method
The emergence of SARS-CoV-2 has prompted scientists to develop viable diagnostic assays that can accurately detect the presence of the virus from biologically derived samples. The antigen–antibody approach, as the second standard approach, revolves around the binding between a synthesized recombinant antigen (produced in the laboratory which mimics specific structures of SARS-CoV-2) and the antibodies present in the body, which elicit an antigen–antibody reaction. Unlike the RT-PCR, the specificity of antigen–antibody testing approach relies on the affinity of target antigen designed. Therefore, designing an antigen specific to the antibodies produced as a result of the present of the virus is crucial for increasing specificity and minimizing the probability of false positive results [42][43].
3.1.3. Antigen-Based Method
The COVID-19 antigen test is another popular diagnosis approach, used as an alternative to the RT-PCR approach. This test is mostly used for early detection of the disease and determining if a patient is contagious. COVID-19 antigen testing revolves around the use of different type samples, which include oral, nasal, and respiratory tracts. The advantages of this approach are that it is very easy to operate, and that it can be used for early detection (i.e., 2 days before the onset of the symptoms). Antigen-based method can be divided into lateral flow rapid-test cassette format and enzyme-linked immunosorbent assay (ELISA) [43][44].
The lateral flow assay-based COVID-19 antigen test, also known as the antigen rapid test, revolves around the collection of plasma, serum, or blood bleed from the fingertip of suspected person, which is subsequently transferred to the test cassette. The test lasts for 20 min, after which the result is displayed. Compared to the lateral flow assay, ELISA-based COVID-19 antigen tests produce accurate and more reliable results. One of the disadvantages of this approach is the requirement of an intensive laboratory procedure, which can be challenging for remote areas with limited healthcare resources [43][45].
3.2. Strengths and Weakness of Molecular Testing
Currently, there are several techniques developed for the detection of pathogens (such as viruses and bacteria). Among these techniques, molecular testing is the most prepared approach, due to its high sensitivity and specificity. These molecular tests include antibody, antigen, and RT-PCR, which can also be subdivided into the molecular test and the serological or antibody test. Molecular testing revolves around the detection of viral RNA in the human body while the virus is still replicating, while antibody assays detect the presence of antibodies produced as a result of human immune response against the virus [7]. In terms of specificity, the antibody assay has a higher specificity, as it can detect if a patient has had COVID-19. One of the limitations of this approach is that antibodies may not be detectable until 1–3 weeks after infection. The antigen test is one of the most rapid and simple technique that can be used to detect SARS-CoV-2 in both asymptomatic and symptomatic patients. Just like the antibody test, the virus can be detected between 5 and 12 days after contact, or after onset of symptoms, and results can be generated after 15 min [46][47]. Despite the test being rapid, fast, and simple, it is hindered by several challenges, which include low sensitivity compared to RT-PCR and the likelihood of false negative results [48].
RT-PCR is the most preferred approach, as it can detect the presence of the virus in an early stage of infection. Even though this test is regarded as the standard approach, one of its limitations is the likelihood of false negative results. Thus, repeated testing is required to overcome this challenge, which increases testing time and cost. Quantitative RT-PCR (RT-qPCR) is another highly sensitive molecular method that can be employed for the detection of COVID-19. The method revolves around the combination of real-time quantification and RT-PCR using fluorescent probes. However, this technique is less widely use due to its expensive instrumentation [49][50].
3.3. Application of Biosensors for the Detection of SARS-CoV-2
The application of a rapid, ultra-sensitive, and quantitative electrochemical biosensor for the detection of SARS-CoV-2 was proposed by Alateef et al. [51]. The biosensor is designed using gold NPs (AUNPs) which is capped with highly specific antisense oligonucleotide, while the sensing probe was immobilized on a paper-based electrochemical device. The sensing mechanism revolves around the interaction between the antisense SSDNA and sensing probe, which generate readout results that can be seen on a hand-held reader. In order to analyze the sensing viability of the paper-based electrochemical biosensor, the developed platform was tested using clinical and vero cells infected with the virus. The performance evaluation of the biosensor resulted in sensitivity of 231 (copies µL−1)−1 and 6.9 copies/UL LOD. Subsequent testing of the device using both samples obtained from 22 patients tested with SARS-CoV-2 confirmed using RT-PCR and 26 healthy patients resulted in 100 accuracy, sensitivity and specificity.
The development of a cheap CRISPR-based POC testing platform known as miSHERLOCK for the detection of SARS-CoV-2 was proposed by de Puig et al. [52]. The sensing mechanism revolves around the collection of unprocessed saliva, followed by extraction, purification, concentration, amplification, and detection based on the interaction between viral NA and guide RNA binds with Cas12a to produce fluorescent visual output within 1 h. The performance of the platform resulted in highly sensitive and multiplexed detection of the virus and mutations associated with two different variants, leading to different LOD in cp/mL. Another distinction of this approach is the application of adjunct smartphones to enable quantification of output, automated interpretation, and the prospect of remote and distributed result reporting.
Song et al. [53] developed an antifouling electrochemical biosensor for the detection of SARS-CoV-2 NA. The nanobiosensor is designed based on electropolymerized polyaniline nanowires and a synthesized Y-shaped peptide which poses antifouling properties. The mechanism behind the working principle of the biosensor revolves around the interaction between immobilized biotin-labeled probes and COVID-19 NA. Evaluation of the performance of the genosensor led to a 3.5 fM detection limit and a wide linear range of 10–14 to 10–9 M.
Detection of SARS-CoV-2 from clinical samples using a field-effect transistor (FET)-biosensor was proposed by Seo et al. [54]. The biosensor was constructed by coating a graphene sheet of the FET with a specific antibody against the viral spike protein. In order to test the viability of the immunobiosensor, several samples, such as antigen proteins, cultured virus, and nasopharyngeal swab samples collected from patients suffering from COVID-19 pneumonia. The performance of the FET-based biosensor for the detection of SARS-CoV-2 spike protein yielded 100 fg/mL concentration in clinical transport medium and 1 fg/mL concentration in phosphate buffer saline. The biosensor was able to detect SARS-CoV-2 in cultured medium with 1.6 × 101 pfu/mL LOD and clinical samples with 2.42 × 102 copies/ML.
Tian et al. [55] developed an electrochemical aptamer-based biosensor for the detection of COVID-19. The biosensor was constructed using metal-organic frameworks MIL-53(AI) which is decorated using AU@Pt NPs and enzymes. The surface of the electrodes was immobilized with dual aptamer as biorecognition element. SARS-CoV-2 is detected based on the interaction between immobilized 2 thiol-modified aptamers (N48 and N61) and SARS-CoV-2 nucleocapsid via the co-catalysis of the nanomaterials, G-quadruplex DNAzyme, and Horseradish Peroxidase (HRP). Evaluation of the biosensor demonstrated 8.33 pg mL−1 LOD and a wide linear range of 0.025 to 50 ng ML−1.
Buyuksunetci et al. [56] developed an electrochemical biosensor for the detection of SARS-CoV-2. The device was constructed using a gold screen printed electrode (AuSPE) and subsequently immobilized with either angiotensin-converting enzyme 2 or CD147. The biosensor is designed based on the interaction between the spike protein and receptors such (ACE2) or CD147. Evaluation of the performance of the biosensor yielded 29,930 ng ML−1 LOD and a linear detection range of 700 ng ML−1 to 1500 ng ML−1, and 1500 ng ML−1 to 7000 ng ML−1 for the detection of spike protein using ACE2, while detection of spike protein using CD147 yielded 38.99 ng ML−1 LOD and linear detection ranges of 500 ng ML−1 to 5000 ng ML−1. The biosensor was also evaluated using clinical samples confirmed with RT-PCR method.
The development of a multiplexed grating-couple fluorescent plasmonic biosensor for the detection of SARS-CoV-2 using either a dried blood spot sample or human blood serum was proposed by Cady et al. [57]. Detection of COVID-19 relied upon the interaction between antibody (IgG) and antigen (nucleocapsid protein, Spike S1 and Spike S1 S2). The performance evaluation of the immunobiosensor produced linear response for serum samples diluted to 1:1600 dilution. The biosensor was also compared with two commercial COVID antibody testing kits (which include Luminex-based microsphere immunoassay and ELISA), which resulted in 100% correlation. Moreover, 63 samples of dried blood spots were tested using the constructed immunobiosensor, which yielded 86.7% sensitivity, and 100% selectivity for detection prior to COVID-19 infection.
Kim et al. [58] developed a sensitive electrochemical biosensor for point-of-care detection of COVID-19. The genobiosensor was designed using a multi-microelectrode array and relied upon the interaction between probes and target genes (N gene and RdRP gene) amplified using Recombinase Polymerase Amplification (RPA) and subsequently detected using pulse voltammetry. This process involved hybridization between thiol-modified primers immobilized on the surface of WE and RPA amplicon, which resulted in reduction of current density due to accumulation of amplicons. The performance of the assay yielded 3925 fg/µL LOD for N gene and 0.972 fg/µL for RdRP gene.
This entry is adapted from the peer-reviewed paper 10.3390/s23010426
References
- Güner, H.R.; Hasanoğlu, İ.; Aktaş, F. COVID-19: Prevention and control measures in community. Turk. J. Med. Sci. 2020, 50, 571–577.
- Ai, T.; Yang, Z.; Hou, H.; Zhan, C.; Chen, C.; Lv, W.; Tao, Q.; Sun, Z.; Xia, L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: A report of 1014 cases. Rad 2020, 296, E32–E40.
- Hamza, A.; Khan, M.A.; Wang, S.H.; Alhaisoni, M.; Alharbi, M.; Hussein, H.S.; Alshazly, H.; Kim, Y.J.; Cha, J. COVID-19 classification using chest X-ray images based on fusion-assisted deep Bayesian optimization and Grad-CAM visualization. Front. Public Health 2022, 10, 1046296.
- Khan, M.A.; Alhaisoni, M.; Nazir, M.; Alqahtani, A.; Binbusayyis, A.; Alsubai, S.; Nam, Y.; Kang, B.G. A Healthcare System for COVID19 Classification Using Multi-Type Classical Features Selection. Comput. Mater. Contin. 2023, 74, 1393–1412.
- Arun, K.R.; Elizabeth, T.R.; Sukumaran, A.; Paul, J.K.; Vasudevan, D.M. COVID-19: Current trends in invitro diagnostics. Indian J. Clin. Biochem. 2020, 35, 285–289.
- Yüce, M.; Filiztekin, E.; Özkaya, K.G. COVID-19 diagnosis—A review of current methods. Biosens. Bioelectron. 2021, 172, 112752.
- Afzal, A. Molecular diagnostic technologies for COVID-19: Limitations and challenges. J. Adv. Res. 2020, 26, 149–159.
- Liu, G.; Rusling, J.F. COVID-19 antibody tests and their limitations. ACS Sens. 2021, 6, 593–612.
- Quraishi, M.; Upadhyay, S.K.; Nigam, A. COVID-19 Diagnostics: A Panoramic View on Its Present Scenario, Challenges and Solutions. India Sect. B Biol. Sci. 2022, 92, 709–721.
- Doi, K. Computer-aided diagnosis in medical imaging: Historical review, current status and future potential. Comput. Med. Imaging Graph. 2007, 31, 198–211.
- Van Ginneken, B.; Schaefer-Prokop, C.M.; Prokop, M. Computer-aided diagnosis: How to move from the laboratory to the clinic. Rad 2011, 261, 719–732.
- Yassin, N.I.; Omran, S.; El Houby, E.M.; Allam, H. Machine learning techniques for breast cancer computer aided diagnosis using different image modalities: A systematic review. Comput. Methods Programs Biomed. 2018, 156, 25–45.
- Ahmad, O.F.; Soares, A.S.; Mazomenos, E.; Brandao, P.; Vega, R.; Seward, E.; Stoyanov, D.; Chand, M.; Lovat, L.B. Artificial intelligence and computer-aided diagnosis in colonoscopy: Current evidence and future directions. Lancet Gastroenterol. Hepatol. 2019, 4, 71–80.
- Litjens, G.; Debats, O.; Barentsz, J.; Karssemeijer, N.; Huisman, H. Computer-aided detection of prostate cancer in MRI. IEEE Trans. Med. Imaging 2014, 33, 1083–1092.
- Iqbal, S.; Ghani Khan, M.U.; Saba, T.; Mehmood, Z.; Javaid, N.; Rehman, A.; Abbasi, R. Deep learning model integrating features and novel classifiers fusion for brain tumor segmentation. Microsc. Res. Tech. 2019, 82, 1302–1315.
- Ibrahim, A.U.; Al-Turjman, F.; Ozsoz, M.; Serte, S. Computer aided detection of tuberculosis using two classifiers. Biomed. Eng./Biomed. Tech. 2022, 67, 513–524.
- Umar Ibrahim, A.; Ozsoz, M.; Serte, S.; Al-Turjman, F.; Habeeb Kolapo, S. Convolutional neural network for diagnosis of viral pneumonia and COVID-19 alike diseases. Expert Syst. 2022, 39, e12705.
- Ibrahim, A.U.; Ozsoz, M.; Serte, S.; Al-Turjman, F.; Yakoi, P.S. Pneumonia classification using deep learning from chest X-ray images during COVID-19. Cognit. Comput. 2021, 1, 1–3.
- Arshad, M.; Khan, M.A.; Tariq, U.; Armghan, A.; Alenezi, F.; Younus Javed, M.; Aslam, S.M.; Kadry, S. A computer-aided diagnosis system using deep learning for multiclass skin lesion classification. Comput. Intell. Neurosci. 2021, 2021, 9619079.
- Razdan, S.; Sharma, S. Internet of Medical Things (IoMT): Overview, emerging technologies, and case studies. IETE Tech. Rev. 2022, 39, 775–788.
- Jeba Kumar, R.J.; Roopa Jayasingh, J.; Telagathoti, D.B. Intelligent Transit Healthcare Schema Using Internet of Medical Things (IoMT) Technology for Remote Patient Monitoring. In Internet of Medical Things; Springer: Cham, Switzerland, 2021; pp. 17–33.
- Dwivedi, R.; Mehrotra, D.; Chandra, S. Potential of Internet of Medical Things (IoMT) applications in building a smart healthcare system: A systematic review. J. Oral Biol. Craniofacial Res. 2021, 12, 302–318.
- Jain, S.; Nehra, M.; Kumar, R.; Dilbaghi, N.; Hu, T.; Kumar, S.; Kaushik, A.; Li, C.Z. Internet of medical things (IoMT)-integrated biosensors for point-of-care testing of infectious diseases. Biosens. Bioelectron. 2021, 179, 113074.
- Manickam, P.; Mariappan, S.A.; Murugesan, S.M.; Hansda, S.; Kaushik, A.; Shinde, R.; Thipperudraswamy, S.P. Artificial intelligence (AI) and internet of medical things (IoMT) assisted biomedical systems for intelligent healthcare. Biosens 2022, 12, 562.
- Ciotti, M.; Ciccozzi, M.; Terrinoni, A.; Jiang, W.C.; Wang, C.B.; Bernardini, S. The COVID-19 pandemic. Crit. Rev. Clin. Lab. Sci. 2020, 57, 365–388.
- Nicholls, J.; Dong, X.P.; Jiang, G.; Peiris, M. SARS: Clinical virology and pathogenesis. Respirology 2003, 8, S6–S8.
- Watanabe, T.; Bartrand, T.A.; Weir, M.H.; Omura, T.; Haas, C.N. Development of a dose-response model for SARS coronavirus. Risk Anal. Int. J. 2010, 30, 1129–1138.
- Oboho, I.K.; Tomczyk, S.M.; Al-Asmari, A.M.; Banjar, A.A.; Al-Mugti, H.; Aloraini, M.S.; Alkhaldi, K.Z.; Almohammadi, E.L.; Alraddadi, B.M.; Gerber, S.I.; et al. 2014 MERS-CoV outbreak in Jeddah—A link to health care facilities. N. Engl. J. Med. 2015, 372, 846–854.
- Ahmadzadeh, J.; Mobaraki, K.; Mousavi, S.J.; Aghazadeh-Attari, J.; Mirza-Aghazadeh-Attari, M.; Mohebbi, I. The risk factors associated with MERS-CoV patient fatality: A global survey. Diagn. Microbiol. Infect. Dis. 2020, 96, 114876.
- Majra, D.; Benson, J.; Pitts, J.; Stebbing, J. SARS-CoV-2 (COVID-19) superspreader events. J. Infect. 2021, 82, 36–40.
- Johansson, M.A.; Quandelacy, T.M.; Kada, S.; Prasad, P.V.; Steele, M.; Brooks, J.T.; Slayton, R.B.; Biggerstaff, M.; Butler, J.C. SARS-CoV-2 transmission from people without COVID-19 symptoms. JAMA Netw. Open 2021, 4, e2035057.
- Uddin, M.; Mustafa, F.; Rizvi, T.A.; Loney, T.; Al Suwaidi, H.; Al-Marzouqi, A.H.; Kamal Eldin, A.; Alsabeeha, N.; Adrian, T.E.; Stefanini, C.; et al. SARS-CoV-2/COVID-19: Viral genomics, epidemiology, vaccines, and therapeutic interventions. Viruses 2020, 12, 526.
- Falzone, L.; Gattuso, G.; Tsatsakis, A.; Spandidos, D.A.; Libra, M. Current and innovative methods for the diagnosis of COVID 19 infection. Int. J. Mol. Med. 2021, 47, 1–23.
- Touma, M. COVID-19: Molecular diagnostics overview. J. Mol. Med. 2020, 98, 947–954.
- Ye, G.; Lin, H.; Chen, S.; Wang, S.; Zeng, Z.; Wang, W.; Zhang, S.; Rebmann, T.; Li, Y.; Pan, Z.; et al. Environmental contamination of SARS-CoV-2 in healthcare premises. J. Infect. 2020, 81, e1–e5.
- Lee, E.Y.; Ng, M.Y.; Khong, P.L. COVID-19 pneumonia: What has CT taught us? Lancet Infect. Dis. 2020, 20, 384–385.
- Lai, C.K.; Lam, W. Laboratory testing for the diagnosis of COVID-19. Biochem. Biophys. Res. Commun. 2021, 538, 226–230.
- Shyu, D.; Dorroh, J.; Holtmeyer, C.; Ritter, D.; Upendran, A.; Kannan, R.; Dandachi, D.; Rojas-Moreno, C.; Whitt, S.P.; Regunath, H. Laboratory tests for COVID-19: A review of peer-reviewed publications and implications for clinical use. Missouri Med. 2020, 117, 184.
- Nyaruaba, R.; Mwaliko, C.; Hong, W.; Amoth, P.; Wei, H. SARS-CoV-2/COVID-19 laboratory biosafety practices and current molecular diagnostic tools. J. Biosaf. Biosecur. 2021, 3, 131–140.
- Smyrlaki, I.; Ekman, M.; Lentini, A.; Rufino de Sousa, N.; Papanicolaou, N.; Vondracek, M.; Aarum, J.; Safari, H.; Muradrasoli, S.; Rothfuchs, A.G.; et al. Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-PCR. Nat. Commun. 2020, 11, 4812.
- Barza, R.; Patel, P.; Sabatini, L.; Singh, K. Use of a simplified sample processing step without RNA extraction for direct SARS-CoV-2 RT-PCR detection. J. Clin. Virol. 2020, 132, 104587.
- Augustine, R.; Das, S.; Hasan, A.; Abdul Salam, S.; Augustine, P.; Dalvi, Y.B.; Varghese, R.; Primavera, R.; Yassine, H.M.; Thakor, A.S.; et al. Rapid antibody-based COVID-19 mass surveillance: Relevance, challenges, and prospects in a pandemic and post-pandemic world. J. Clin. Med. 2020, 9, 3372.
- La Marca, A.; Capuzzo, M.; Paglia, T.; Roli, L.; Trenti, T.; Nelson, S.M. Testing for SARS-CoV-2 (COVID-19): A systematic review and clinical guide to molecular and serological in-vitro diagnostic assays. Reprod. Biomed. Online 2020, 41, 483–499.
- Lv, Y.; Ma, Y.; Si, Y.; Zhu, X.; Zhang, L.; Feng, H.; Tian, D.; Liao, Y.; Liu, T.; Lu, H.; et al. Rapid SARS-CoV-2 antigen detection potentiates early diagnosis of COVID-19 disease. Biosci. Trends 2021, 15, 93–99.
- Li, K.; Tong, C.; Ha, X.; Zeng, C.; Chen, X.; Xu, F.; Yang, J.; Du, H.; Chen, Y.; Cai, J.; et al. Development and clinical evaluation of a rapid antibody lateral flow assay for the diagnosis of SARS-CoV-2 infection. BMC Infect. Dis. 2021, 21, 860.
- Kyosei, Y.; Namba, M.; Yamura, S.; Takeuchi, R.; Aoki, N.; Nakaishi, K.; Watabe, S.; Ito, E. Proposal of de novo antigen test for COVID-19: Ultrasensitive detection of spike proteins of SARS-CoV-2. Diagnostics 2020, 10, 594.
- Pray, I.W. Performance of an antigen-based test for asymptomatic and symptomatic SARS-CoV-2 testing at two university campuses—Wisconsin, September–October 2020. Morb. Mortal. Wkly. Rep. 2021, 69, 1642–1647.
- Jayamohan, H.; Lambert, C.J.; Sant, H.J.; Jafek, A.; Patel, D.; Feng, H.; Beeman, M.; Mahmood, T.; Nze, U.; Gale, B.K. SARS-CoV-2 pandemic: A review of molecular diagnostic tools including sample collection and commercial response with associated advantages and limitations. Anal. Bioanal. Chem. 2021, 413, 49–71.
- Surkova, E.; Nikolayevskyy, V.; Drobniewski, F. False-positive COVID-19 results: Hidden problems and costs. Lancet Respir. Med. 2020, 8, 1167–1168.
- Dao Thi, V.L.; Herbst, K.; Boerner, K.; Meurer, M.; Kremer, L.P.; Kirrmaier, D.; Freistaedter, A.; Papagiannidis, D.; Galmozzi, C.; Stanifer, M.L.; et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020, 12, eabc7075.
- Alafeef, M.; Dighe, K.; Moitra, P.; Pan, D. Rapid, ultrasensitive, and quantitative detection of SARS-CoV-2 using antisense oligonucleotides directed electrochemical biosensor chip. ACS Nano 2020, 14, 17028–17045.
- de Puig, H.; Lee, R.A.; Najjar, D.; Tan, X.; Soenksen, L.R.; Angenent-Mari, N.M.; Donghia, N.M.; Weckman, N.E.; Ory, A.; Ng, C.F.; et al. Minimally instrumented SHERLOCK (miSHERLOCK) for CRISPR-based point-of-care diagnosis of SARS-CoV-2 and emerging variants. Sci. Adv. 2021, 7, eabh2944.
- Song, Z.; Ma, Y.; Chen, M.; Ambrosi, A.; Ding, C.; Luo, X. Electrochemical biosensor with enhanced antifouling capability for COVID-19 nucleic acid detection in complex biological media. Anal. Chem. 2021, 93, 5963–5971.
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.H.; Choi, M.; Ku, K.B.; Lee, C.S.; Jun, S.; Park, D.; Kim, H.G.; et al. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano 2020, 14, 5135–5142.
- Tian, J.; Liang, Z.; Hu, O.; He, Q.; Sun, D.; Chen, Z. An electrochemical dual-aptamer biosensor based on metal-organic frameworks MIL-53 decorated with Pt nanoparticles and enzymes for detection of COVID-19 nucleocapsid protein. Electrochim. Acta 2021, 387, 138553.
- Büyüksünetçi, Y.T.; Çitil, B.E.; Anık, Ü. An impedimetric approach for COVID-19 detection. Analyst 2022, 147, 130–138.
- Cady, N.C.; Tokranova, N.; Minor, A.; Nikvand, N.; Strle, K.; Lee, W.T.; Page, W.; Guignon, E.; Pilar, A.; Gibson, G.N. Multiplexed detection and quantification of human antibody response to COVID-19 infection using a plasmon enhanced biosensor platform. Biosens. Bioelectron. 2021, 171, 112679.
- Kim, H.E.; Schuck, A.; Lee, S.H.; Lee, Y.; Kang, M.; Kim, Y.S. Sensitive electrochemical biosensor combined with isothermal amplification for point-of-care COVID-19 tests. Biosens. Bioelectron. 2021, 182, 113168.
This entry is offline, you can click here to edit this entry!
