Chemoresistance and metastasis are the main malignant features in cancer, resulting the unfavorable outcome. Accumulating evidence suggest dyregulated autophagy contributes to resistance to conventional therapy and metastasis which can be regulated by lncRNAs, circRNAs and miRNAs. Autophagy consists of a cascade of steps controlled by different autophagy related genes that can be regulated those ncRNAs. The lncRNAs, circRNAs, miRNAs, mRNAs compromised competing endogenous RNA network and participate in carcinogenesis. Here, we attempt to review the role of ceRNAs in cancer metastasis and chemoresistance through autophagy regulation.
- autophag y
- ceRNA
- cancer
1. Introduction
Autophagy is an evolutionarily conserved system to maintain homeostasis in various cells. In general, there are three main types of autophagy: macroautophagy, microautophagy and mitoautophagy [1]. Mitoautophagy can remove damaged mitochondria specifically by autophagosomes for lysosomal degradation while microautophagy, mainly in plants and fungi, may complete the isolation and uptake of cell components by directly enveloping them with the vacuolar/lysosomal membrane. Macroautophagy is the best-characterized process that aims to remove unwanted or damaged organelles and aggregated proteins by lysosome degradation. In the narrow sense, some scientists (including those in this article) have referred to macroautophagy as autophagy.
The dysregulation of autophagy has been reported in many human diseases, such as infection, cardiovascular and neurodegenerative diseases, and cancers [2]. Metastasis and chemoresistance are two major factors that are associated with cancer recurrence and dismal clinical outcomes. Moreover, metastasis and chemoresistance are closely linked in cancers, as metastatic cancer cells have a propensity for chemoresistance [3][4]. The roles of autophagy in the processes of cancer metastasis and chemoresistance are starting to be recognized. Autophagy is critically involved in many aspects of cancer metastasis, including epithelial–mesenchymal transition (EMT), invasion, migration, anoikis resistance, crosstalk between cancer and stromal cells, and immune suppression [5]. Cancer cells may survive chemotherapy through protective autophagy [6][7][8].
Long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) are non-coding RNAs (ncRNAs) with respective lengths of more than 200 nucleotides and about 22 nucleotides. Both participate in various physiological and pathological processes involving tumorigenesis, such as cell proliferation, apoptosis, EMT, invasion, migration, chemoresistance, and stemness [9]. lncRNAs can regulate gene expression through repressing chromatin components [10], mediating epigenetic silencing, interacting with Polycomb complex, and modulating transcription activation [11]. Considering that there is a complex interplay among messenger RNA (mRNA) and ncRNAs [12][13], lncRNAs also function as miRNA sponges by competitively binding with miRNAs, thereby releasing the targeted mRNAs [14][15]. These lncRNAs are called competing endogenous RNAs (ceRNAs). They can decrease the activity or expression of miRNAs [16][17]. Circular RNA (circRNA) is another kind of ncRNA comprising covalently closed single-stranded loops. Recent advances in RNA sequencing and bioinformatics tools have led to the discovery and identification of thousands of circRNAs and validated their important roles in the pathogenesis of cancers [18][19][20][21]. They can mainly function as ceRNAs to modulate gene transcription by interacting with miRNAs or lncRNAs, sponging miRNAs, or RNA-binding proteins, and rarely can be translated into proteins [22][23][24][25]. In addition to lncRNA and circRNA, other ceRNAs have also been identified by bioinformatics analysis and experimental evidence, such as pseudogenes, mRNA including those expressing 3′-untranslated regions, virus non-coding RNAs, and genomic viral RNAs [26]. Pseudogenes, the relicts of parental gene that are unable to encode full-length proteins, can regulate the expression of parental genes via ceRNA networks, and participate in tumorigenesis [27]. Some mRNAs can function as ceRNAs and can be associated with human diseases such as cardiovascular diseases and glioblastoma multiforme [28][29]. Accumulative data have indicated that ceRNAs, particularly lncRNAs and circRNAs, can bridge the interplay between autophagy and chemoresistance or metastasis in cancers [30][31][32][33][34].
2. An Overview of Autophagy-Regulated Cancer Metastasis and Chemoresistance
Emerging evidence has supported the roles of autophagy in the processes of cancer metastasis and chemoresistance. Cancer cells may succeed in metastasis through tumor-promoting autophagy [5], and survival against chemotherapy through protective autophagy [6][7][8].
In most cancers, the activation of autophagy promotes chemoresistance and indicates poor survival. Chemotherapeutic agents induce stress, thus activating autophagy as a cellular adaptive response [35][36]. Recently, it has been demonstrated that DNA damage can induce the process of autophagy and upregulate related genes, essentially resulting in chemoresistance [37]. Therefore, the activation of autophagy becomes a novel molecular mechanism for chemoresistance modulated by some lncRNAs in cancer cells, such as GBCDRlnc1 in gallbladder cancer [38] and MALAT1 in gastric cancer [39]. Under some circumstances, autophagy inhibition may restore chemotherapy sensitivity. For instance, autophagy suppression by Atg7 knockdown enhanced chemosensitivity and prolonged overall survival in AML mouse models [35]. Autophagy mediated by OPN/NF-κB signaling is essential for pancreatic cancer stem cells, while pharmacological inhibition of autophagy increased drug sensitivity [40]. These findings suggest a link between autophagy and cancer stem cells—the root cause for chemoresistance and disease relapse. Additionally, autophagy suppression can enhance chemotherapy response in triple-negative breast cancers (TNBCs) [36]. In HCC cells, miR-541 increased their sensitivity to sorafenib by inhibiting autophagy [41]. Moreover, N6-isopentenyladenosine also improved chemotherapy sensitivity by impairing autophagy in melanoma [42].
Cancer cells experience invasion into the circulation, migration into the pre-metastatic niche, and colonization at the new place [43]. A substantial body of evidence has shown that autophagy is crucial for cancer metastasis through regulating many important steps in this process, such as tumor invasion, migration, EMT, the crosstalk between stromal cells and tumor cells, and immune surveillance [43][44][45][46]. The precision function of autophagy in cancer metastasis remains controversial to date, with tumor-promoting roles in most studies and suppressive roles in a few [47]. A high level of ATG5 is required for metastasis in the mouse models of pancreatic ductal adenocarcinoma [48], while elevated LC3B has been indicated to be associated with invasion and metastasis in solid tumors [49]. Ube2v1, a ubiquitin-conjugating E2 enzyme variant, can facilitate metastasis in colorectal cancer by inhibiting autophagy. Importantly, rapamycin and trehalose treatment may reverse Ube2v1-mediated metastasis in mouse models [50]. In ovarian carcinoma, circMUC16-mediated autophagy promotes metastasis by directly binding to ATG13 and enhancing its expression. Moreover, cirMUC16 could also function as a ceRNA for miR-199a-5p and relieve its inhibitory on Beclin-1 and RUNX1 [51]. Nevertheless, in breast cancer, the expression of autophagy-related genes negatively correlates with pre-metastasis signatures, and thereof restricts tumor metastasis [52][53].
3. Conclusion and Future Perspectives
In conclusion, ceRNAs play important roles in many human cancers. Most ceRNAs positively regulate autophagy unless they sponge miRNAs targeting mTOR. Nearly all ceRNAs function as oncogenes, with rare exceptions (Table 1).
Most metastatic cancer cells are chemoresistant [3][54]. It is prerequisite to explore the underlying molecular link between metastasis and chemotherapy. Many factors significantly contribute to drug resistance, such as cancer cell heterogeneity, drug efflux, strength of DNA repair, cancer cell stemness, and dysregulated proliferation/apoptosis pathways [55][56][57]. Under unfavorable circumstances, autophagy activation can protect cells from stress-induced injury [58][59]. Unequivocally, chemotherapy drugs can activate autophagy in cancer cells [60]. The metastasis of cancer harbors some key events, such as EMT, invasion into vessels, resistance to anoikis in circulation, and travelling to the secondary site [45]. During this process, cancer cells struggle to survive against various stresses. Likewise, autophagy also plays an important part in metastasis against these stresses [61]. Moreover, even in the secondary site, it is important to build up an appropriate microenvironment—that is, the pre-metastasis niche [62][63]. Autophagy is essential for niche formation owing to its biological effects on stromal cells, immune escape, angiogenesis, and lymphangiogenesis [62]. Therefore, further clarification of the mechanism of autophagy regulation in metastasis and chemoresistance may greatly improve the efficacy of chemotherapy and survival in cancer patients.
lncRNAs, circRNAs, and miRNAs are critical in tumorigenesis and may serve as promising therapeutic targets and biomarkers for cancers [9][64][65]. ceRNAs communicate with different types of RNAs in cancer progression, forming the network among lncRNAs, circRNAs, miRNAs, and mRNAs [15][66]. In this manuscript, we summarized the role of ceRNA-regulated autophagy in chemoresistance and metastasis. ceRNAs can regulate the expression of autophagy-related genes via binding with some miRNAs. These genes include mTOR, ULK1, ATG3, ATG4, ATG5, ATG7, ATG12, ATG13, and ATG14, which are critical for autophagosome initiation, elongation, and fusion with lysosomes (Figure 1).
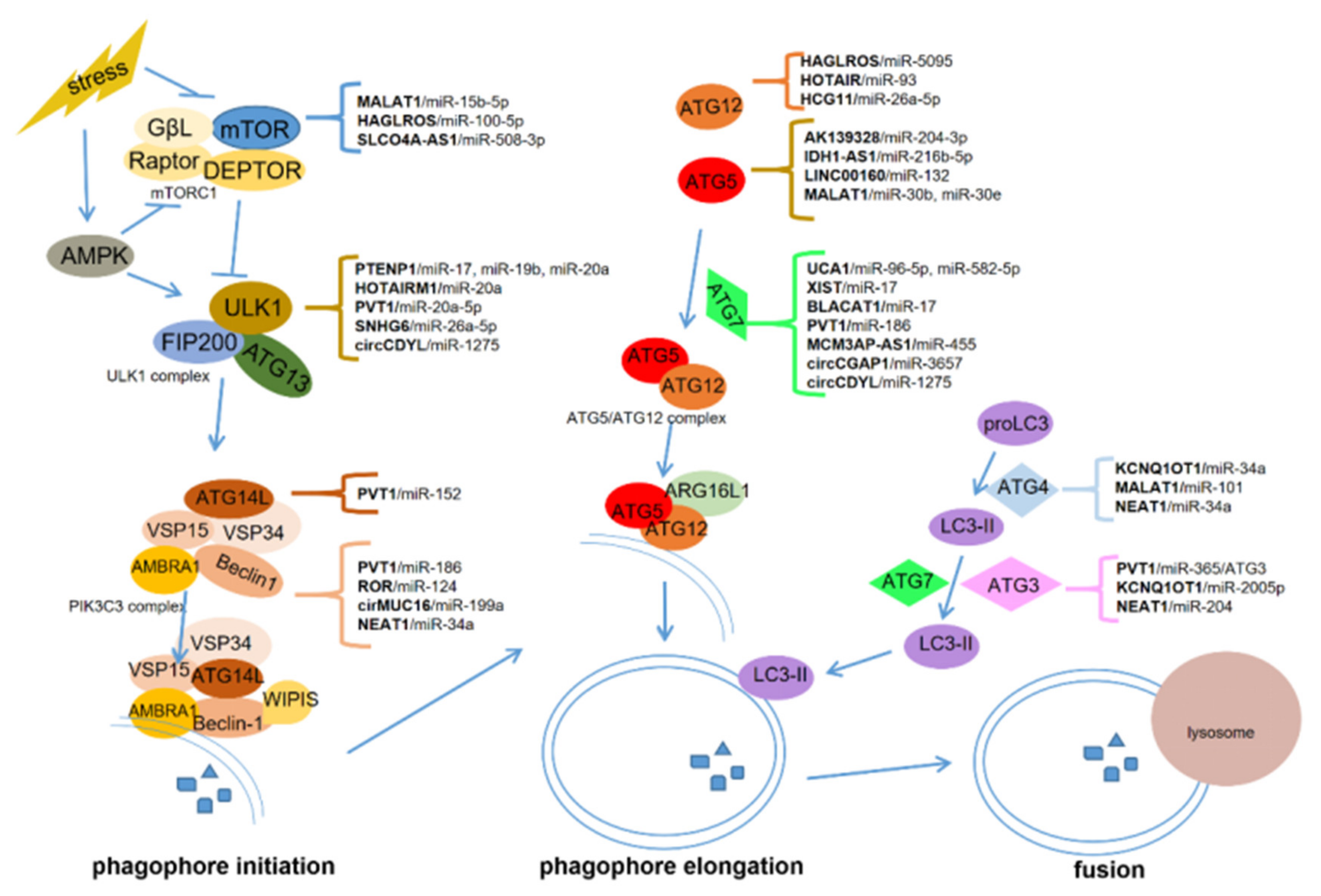
To date, ceRNA-mediated autophagy is mostly known as a positive regulator of chemoresistance and metastasis. These ceRNAs can promote chemoresistance through proliferation inhibition and apoptosis induction, and facilitate metastasis by inducing EMT, invasion, migration, angiogenesis, and modulating the pre-metastasis niche. Some ceRNAs can be packaged in exosomes which are derived from cancer cells or bone marrow mesenchymal stem cells. The exosomal ceRNAs contribute to optimizing the microenvironment for cancer metastasis. However, more work is required to uncover how ceRNA-mediated autophagy regulates chemoresistance and metastasis in cancers. Particular issues might be focused on the potential role of ceRNAs in the crosstalk among cancer cells and the tumor microenvironment composed of stromal cells, endothelial cells (angiogenesis), and immunocytes. Nevertheless, several critical issues should first be addressed.
The ceRNA theory is supported by a great body of experimental data on lncRNAs and circRNAs. The studies are invariably associated with ectopic vectors which produce much higher than physiological levels in cancer cells, and unavoidably present non-physiological phenotypes [67]. The stoichiometry required for a ceRNA, a competitor for miRNAs, includes a substantially higher endogenous content and affinity with linear transcripts over miRNAs; therefore, it is very unlikely that a low-level ceRNA or physiological changes in the expression of an individual ceRNA would effectively regulate a far more abundant miRNA target. In fact, the abundance of ceRNAs such as lncRNA and circRNA in cancers has always been a major concern. The absolute levels of functional ceRNAs in cancer tissues have not been well addressed to date, owing to the difficulty in accurately quantifying ceRNAs and due to some technical problems in tissue preparation. The unification of accurate quantification becomes a prerequisite for ceRNA studies in cancers before we can find a functional ceRNA with considerably high contents.
A ceRNA may target multiple miRNAs, and the potential target miRNAs may also have several target genes. It is reasonable to reckon that a subtle alteration of the miRNA by its upstream ceRNA may have a significant effect on the target genes in the process of autophagy. The development of a ceRNA network is crucial for better understanding the molecular mechanisms and potential utilities underlying gene expression regulated by ceRNAs. However, ceRNA networks have so far predominantly been constructed by bioinformatic prediction, with very limited experimental data. Another important issue with regard to the interaction between ceRNA and miRNA is the problem of “degradation” or “inhibition”, which is crucial for ceRNA function [67]. There is still conflicting evidence describing the effect of ceRNA–protein complexes on the degradation of miRNA after binding. It may be determined by the status of the binding sites between circRNAs and miRNAs: “degradation” in the case of completely complementary, or “inhibition” when partially matched [68].
Bioinformatic analysis, dual-luciferase assay, and methods measuring gene expression (e.g., reverse transcription-PCR (qRT-PCR) and Western blot) are the most commonly applied approaches in ceRNA investigations [26]. Results from bioinformatic prediction should be validated by experimental data. Dual-luciferase assay and gene expression analysis are not direct and sound methods to establish the interaction among ceRNA, miRNA, and mRNA. The reliable methods are currently RIP and RNA pull-down. However, both methods have only been applied in a small number of studies. Importantly, the experimental results are obtained from gain- or loss-of-function assays in vitro and, uncommonly, in vivo. The technical limitations and defects should be appreciated in the assessment of the current studies. From the perspective of clinical relevance, future elaborate work in primary-cancer-derived organoids which repeat the pathological features of cancer [69], and studies in vivo will consolidate the interaction among ceRNA, miRNA, and RNA; therefore, they will ultimately provide great insights into carcinogenesis including autophagy-related cancer chemoresistance and metastasis.
This entry is adapted from the peer-reviewed paper 10.3390/cancers12102926
References
- Zhang, H.; Duan, C.; Yang, H. Defective autophagy in Parkinson’s disease: Lessons from genetics. Mol. Neurobiol. 2015, 51, 89–104.
- Jiang, P.; Mizushima, N. Autophagy and human diseases. Cell Res. 2014, 24, 69–79.
- Acharyya, S.; Oskarsson, T.; Vanharanta, S.; Malladi, S.; Kim, J.; Morris, P.G.; Manova-Todorova, K.; Leversha, M.; Hogg, N.; Seshan, V.E.; et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell 2012, 150, 165–178.
- Ren, H.; Du, P.; Ge, Z.; Jin, Y.; Ding, D.; Liu, X.; Zou, Q. TWIST1 and BMI1 in cancer metastasis and chemoresistance. J. Cancer 2016, 7, 1074–1080.
- Dower, C.M.; Wills, C.A.; Frisch, S.M.; Wang, H.-G. Mechanisms and context underlying the role of autophagy in cancer metastasis. Autophagy 2018, 14, 1110–1128.
- Sui, X.; Chen, R.; Wang, Z.; Huang, Z.; Kong, N.; Zhang, M.; Han, W.; Lou, F.; Yang, J.; Zhang, Q.; et al. Autophagy and chemotherapy resistance: A promising therapeutic target for cancer treatment. Cell Death Dis. 2013, 4, e838.
- Yu, T.; Guo, F.; Yu, Y.; Sun, T.; Ma, D.; Han, J.; Qian, Y.; Kryczek, I.; Sun, D.; Nagarsheth, N.; et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 2017, 170, 548–563.e16.
- Pérez-Hernández, M.; Arias, A.; Martínez-García, D.; Pérez-Tomás, R.; Quesada, R.; Soto-Cerrato, V. Targeting autophagy for cancer treatment and tumor chemosensitization. Cancers 2019, 11, 1599.
- Wei, L.; Sun, J.; Zhang, N.; Zheng, Y.; Wang, X.; Lv, L.; Liu, J.; Xu, Y.; Shen, Y.; Yang, M. Noncoding RNAs in gastric cancer: Implications for drug resistance. Mol. Cancer 2020, 19, 62.
- Quinodoz, S.A.; Guttman, M. Long noncoding RNAs: An emerging link between gene regulation and nuclear organization. Trends Cell Biol. 2014, 24, 651–663.
- Bonasio, R.; Shiekhattar, R. Regulation of transcription by long noncoding RNAs. Annu. Rev. Genet. 2014, 48, 433–455.
- Lou, W.; Ding, B.; Fu, P. Pseudogene-derived lncRNAs and their miRNA sponging mechanism in human cancer. Front. Cell Dev. Biol. 2020, 8, 85.
- Cui, X.; Wang, J.; Guo, Z.; Li, M.; Li, M.; Liu, S.; Liu, H.; Li, W.; Yin, X.; Tao, J.; et al. Emerging function and potential diagnostic value of circular RNAs in cancer. Mol. Cancer 2018, 17, 123.
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nat. Cell Biol. 2014, 505, 344–352.
- Karreth, F.A.; Pandolfi, P.P. ceRNA cross-talk in cancer: When ce-bling rivalries go awry. Cancer Discov. 2013, 3, 1113–1121.
- Wang, J.; Liu, X.; Wu, H.; Ni, P.; Gu, Z.; Qiao, Y.; Chen, N.; Sun, F.; Fan, Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010, 38, 5366–5383.
- Li, S.-P.; Xu, H.-X.; Yu, Y.; He, J.-D.; Wang, Z.; Xu, Y.-J.; Wang, C.-Y.; Zhang, H.-M.; Zhang, R.-X.; Zhang, J.-J.; et al. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget 2016, 7, 42431–42446.
- Zhang, Q.; Wang, W.; Zhou, Q.; Chen, C.; Yuan, W.; Liu, J.; Li, X.; Sun, Z. Roles of circRNAs in the tumour microenvironment. Mol. Cancer 2020, 19, 14.
- Shan, C.; Zhang, Y.; Hao, X.; Gao, J.; Chen, X.; Wang, K. Biogenesis, functions and clinical significance of circRNAs in gastric cancer. Mol. Cancer 2019, 18, 136.
- Lei, M.; Zheng, G.; Ning, Q.; Zheng, J.; Dong, D. Translation and functional roles of circular RNAs in human cancer. Mol. Cancer 2020, 19, 30.
- Su, M.; Xiao, Y.; Ma, J.; Tang, Y.; Tian, B.; Zhang, Y.; Li, X.; Wu, Z.; Yang, D.; Zhou, Y.; et al. Circular RNAs in Cancer: Emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol. Cancer 2019, 18, 90.
- Sun, J.; Li, B.; Shu, C.; Ma, Q.; Wang, J. Functions and clinical significance of circular RNAs in glioma. Mol. Cancer 2020, 19, 34.
- Jia, Y.; Li, X.; Nan, A.; Zhang, N.; Chen, L.; Zhou, H.; Zhang, H.; Qiu, M.; Zhu, J.; Ling, Y.; et al. Circular RNA 406961 interacts with ILF2 to regulate PM2.5-induced inflammatory responses in human bronchial epithelial cells via activation of STAT3/JNK pathways. Environ. Int. 2020, 141, 105755.
- Han, D.; Wang, Y.; Wang, Y.; Dai, X.; Zhou, T.; Chen, J.; Tao, B.; Zhang, J.; Cao, F. The tumor-suppressive human circular RNA CircITCH sponges miR-330-5p to ameliorate doxorubicin-induced cardiotoxicity through upregulating SIRT6, Survivin, and SERCA2a. Circ. Res. 2020, 127.
- Chen, L.-L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 2020, 21, 475–490.
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283.
- An, Y.; Furber, K.L.; Ji, S. Pseudogenes regulate parental gene expression via ceRNA network. J. Cell. Mol. Med. 2017, 21, 185–192.
- Song, C.; Zhang, J.; Qi, H.; Feng, C.; Chen, Y.; Cao, Y.; Ba, L.; Ai, B.; Wang, Q.; Huang, W.; et al. The global view of mRNA-related ceRNA cross-talks across cardiovascular diseases. Sci. Rep. 2017, 7, 10185.
- Zhu, X.; Jiang, L.; Yang, H.; Chen, T.; Wu, X.; Lv, K. Analyzing the lncRNA, miRNA, and mRNA-associated ceRNA networks to reveal potential prognostic biomarkers for glioblastoma multiforme. Cancer Cell Int. 2020, 20, 393.
- Sun, T. Long noncoding RNAs act as regulators of autophagy in cancer. Pharmacol. Res. 2018, 129, 151–155.
- Kulkarni, B.; Kirave, P.; Gondaliya, P.; Jash, K.; Jain, A.; Tekade, R.K.; Kalia, K. Exosomal miRNA in chemoresistance, immune evasion, metastasis and progression of cancer. Drug Discov. Today 2019, 24, 2058–2067.
- Huang, F.; Chen, W.; Peng, J.; Li, Y.; Zhuang, Y.; Zhu, Z.; Shao, C.-K.; Yang, W.; Yao, H.; Zhang, S. LncRNA PVT1 triggers Cyto-protective autophagy and promotes pancreatic ductal adenocarcinoma development via the miR-20a-5p/ULK1 Axis. Mol. Cancer 2018, 17, 98.
- Bermúdez, M.; Aguilar-Medina, M.; Lizárraga-Verdugo, E.; Avendaño-Félix, M.; Silva-Benítez, E.; López-Camarillo, C.; Ramos-Payán, R. LncRNAs as regulators of autophagy and drug resistance in colorectal Cancer. Front. Oncol. 2019, 9, 1008.
- Cristóbal, I.; Caramés, C.; Rubio, J.; Sanz-Álvarez, M.; Luque, M.; Madoz-Gúrpide, J.; Rojo, F.; García-Foncillas, J. Functional and clinical impact of CircRNAs in oral cancer. Cancers 2020, 12, 1041.
- Piya, S.; Kornblau, S.M.; Ruvolo, V.; Mu, H.; Ruvolo, P.P.; McQueen, T.; Davis, E.; Hail, N.; Kantarjian, H.; Andreeff, M.; et al. Atg7 suppression enhances chemotherapeutic agent sensitivity and overcomes stroma-mediated chemoresistance in acute myeloid leukemia. Blood 2016, 128, 1260–1269.
- Chittaranjan, S.; Bortnik, S.; Dragowska, W.H.; Xu, J.; Abeysundara, N.; Leung, A.; Go, N.E.; DeVorkin, L.; Weppler, S.A.; Gelmon, K.; et al. Autophagy inhibition augments the anticancer effects of epirubicin treatment in anthracycline-sensitive and -resistant triple-negative breast cancer. Clin. Cancer Res. 2014, 20, 3159–3173.
- Chen, Y.; Wu, J.; Liang, G.; Geng, G.; Zhao, F.; Yin, P.; Nowsheen, S.; Wu, C.; Li, Y.; Li, L.; et al. CHK2-FOXK axis promotes transcriptional control of autophagy programs. Sci. Adv. 2020, 6, eaax5819.
- Cai, Q.; Wang, S.; Jin, L.; Weng, M.; Zhou, D.; Wang, J.; Tang, Z.; Quan, Z. Long non-coding RNA GBCDRlnc1 induces chemoresistance of gallbladder cancer cells by activating autophagy. Mol. Cancer 2019, 18, 82.
- Yiren, H.; Yingcong, Y.; Sunwu, Y.; Keqin, L.; XiaoChun, T.; Senrui, C.; Ende, C.; Xizhou, L.; Yanfan, C. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol. Cancer 2017, 16, 174.
- Yang, M.-C.; Wang, H.-C.; Hou, Y.-C.; Tung, H.-L.; Chiu, T.-J.; Shan, Y.-S. Blockade of autophagy reduces pancreatic cancer stem cell activity and potentiates the tumoricidal effect of gemcitabine. Mol. Cancer 2015, 14, 179.
- Xu, W.-P.; Liu, J.-P.; Feng, J.-F.; Zhu, C.-P.; Yang, Y.; Zhou, W.-P.; Ding, J.; Huang, C.-K.; Cui, Y.-L.; Ding, C.-H.; et al. miR-541 potentiates the response of human hepatocellular carcinoma to sorafenib treatment by inhibiting autophagy. Gut 2019, 69, 1309–1321.
- Ranieri, R.; Ciaglia, E.; Amodio, G.; Picardi, P.; Proto, M.C.; Gazzerro, P.; Laezza, C.; Remondelli, P.; Bifulco, M.; Pisanti, S. N6-isopentenyladenosine dual targeting of AMPK and Rab7 prenylation inhibits melanoma growth through the impairment of autophagic flux. Cell Death Differ. 2018, 25, 353–367.
- Su, Z.; Yang, Z.; Xu, Y.; Chen, Y.; Yu, Q. Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol. Cancer 2015, 14, 48.
- Vera-Ramirez, L. Cell-intrinsic survival signals. The role of autophagy in metastatic dissemination and tumor cell dormancy. Semin. Cancer Biol. 2020, 60, 28–40.
- E Mowers, E.; Sharifi, M.N.; MacLeod, K.F. Autophagy in cancer metastasis. Oncogene 2017, 36, 1619–1630.
- Chen, H.-T.; Liu, H.; Mao, M.-J.; Tan, Y.; Mo, X.-Q.; Meng, X.-J.; Cao, M.-T.; Zhong, C.-Y.; Liu, Y.; Shan, H.; et al. Crosstalk between autophagy and epithelial-mesenchymal transition and its application in cancer therapy. Mol. Cancer 2019, 18, 101.
- Li, X.; He, S.-K.; Ma, B. Autophagy and autophagy-related proteins in cancer. Mol. Cancer 2020, 19, 12.
- Görgülü, K.; Diakopoulos, K.N.; Ai, J.; Schoeps, B.; Kabacaoglu, D.; Karpathaki, A.-F.; Ciecielski, K.J.; Kaya-Aksoy, E.; Ruess, D.A.; Berninger, A.; et al. Levels of the autophagy-related 5 protein affect progression and metastasis of pancreatic tumors in mice. Gastroenterology 2019, 156, 203–217.e20.
- Lazova, R.; Camp, R.L.; Klump, V.; Siddiqui, S.F.; Amaravadi, R.K.; Pawelek, J.M. Punctate LC3B expression is a common feature of solid tumors and associated with proliferation, metastasis, and poor outcome. Clin. Cancer Res. 2011, 18, 370–379.
- Shen, T.; Cai, L.-D.; Liu, Y.-H.; Li, S.; Gan, W.-J.; Li, X.-M.; Wang, J.-R.; Guo, P.-D.; Zhou, Q.; Lu, X.-X.; et al. Ube2v1-mediated ubiquitination and degradation of Sirt1 promotes metastasis of colorectal cancer by epigenetically suppressing autophagy. J. Hematol. Oncol. 2018, 11, 95.
- Gan, X.; Zhu, H.; Jiang, X.; Obiegbusi, S.C.; Yong, M.; Long, X.; Hu, J. CircMUC16 promotes autophagy of epithelial ovarian cancer via interaction with ATG13 and miR-199a. Mol. Cancer 2020, 19, 45.
- Marsh, T.; Kenific, C.M.; Suresh, D.; Gonzalez, H.; Shamir, E.R.; Mei, W.; Tankka, A.; Leidal, A.M.; Kalavacherla, S.; Woo, K.; et al. Autophagic degradation of NBR1 restricts metastatic outgrowth during mammary tumor progression. Dev. Cell 2020, 52, 591–604.e6.
- Mao, L.; Zhan, Y.-Y.; Wu, B.; Yu, Q.; Xu, L.; Hong, X.; Zhong, L.; Mi, P.; Xiao, L.; Wang, X.; et al. ULK1 phosphorylates Exo70 to suppress breast cancer metastasis. Nat. Commun. 2020, 11, 117.
- Ojha, R.; Bhattacharyya, S.; Singh, S.K. Autophagy in cancer stem cells: A potential link between chemoresistance, recurrence, and metastasis. BioRes. Open Access 2015, 4, 97–108.
- E Pollok, K.; Saadatzadeh, R.; Zd, N.; Gj, K.; Sk, G.; Ba, J.; Ia, C.; P, M.; Jn, S.; Ld, S. Faculty Opinions recommendation of DNA repair capacity in multiple pathways predicts chemoresistance in glioblastoma multiforme. Cancer Res. 2017, 77, 198–206.
- Abdullah, L.N.; Chow, E.K. Mechanisms of chemoresistance in cancer stem cells. Clin. Transl. Med. 2013, 2, 3.
- Shi, W.-J.; Gao, J.-B. Molecular mechanisms of chemoresistance in gastric cancer. World J. Gastrointest. Oncol. 2016, 8, 673–681.
- Kroemer, G.; Mariño, G.; Levine, B. Autophagy and the Integrated Stress Response. Mol. Cell 2010, 40, 280–293.
- Ogata, M.; Hino, S.-I.; Saito, A.; Morikawa, K.; Kondo, S.; Kanemoto, S.; Murakami, T.; Taniguchi, M.; Tanii, I.; Yoshinaga, K.; et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell. Biol. 2006, 26, 9220–9231.
- Garbar, C.; Mascaux, C.; Giustiniani, J.; Merrouche, Y.; Bensussan, A. Chemotherapy treatment induces an increase of autophagy in the luminal breast cancer cell MCF7, but not in the triple-negative MDA-MB231. Sci. Rep. 2017, 7, 1–11.
- Peng, Y.-F.; Shi, Y.; Ding, Z.-B.; Ke, A.-W.; Gu, C.-Y.; Hui, B.; Zhou, J.; Qiu, S.-J.; Dai, Z.; Fan, J. Autophagy inhibition suppresses pulmonary metastasis of HCC in mice via impairing anoikis resistance and colonization of HCC cells. Autophagy 2013, 9, 2056–2068.
- Liu, Y.; Cao, X. Characteristics and Significance of the Pre-metastatic Niche. Cancer Cell 2016, 30, 668–681.
- Celià-Terrassa, T.; Kang, Y. Metastatic niche functions and therapeutic opportunities. Nat. Cell Biol. 2018, 20, 868–877.
- Prabhu, K.S.; Raza, A.; Karedath, T.; Raza, S.S.; Fathima, H.; Ahmed, E.I.; Kuttikrishnan, S.; Therachiyil, L.; Kulinski, M.; Dermime, S.; et al. Non-coding RNAs as regulators and markers for targeting of breast cancer and cancer stem cells. Cancers 2020, 351.
- Liu, Y.; Cheng, Z.; Pang, Y.; Cui, L.; Qian, T.; Quan, L.; Zhao, H.; Shi, J.; Ke, X.; Fu, L. Role of microRNAs, circRNAs and long noncoding RNAs in acute myeloid leukemia. J. Hematol. Oncol. 2019, 12, 51.
- Guo, L.-L.; Song, C.-H.; Wang, P.; Dai, L.-P.; Zhang, J.-Y.; Wang, K. Competing endogenous RNA networks and gastric cancer. World J. Gastroenterol. 2015, 21, 11680–11687.
- Li, H.; Ma, X.; Li, H. Intriguing circles: Conflicts and controversies in circular RNA research. Wiley Interdiscip. Rev. RNA 2019, 10, e1538.
- Piwecka, M.; Glažar, P.; Hernandez-Miranda, L.R.; Memczak, S.; Wolf, S.A.; Rybak-Wolf, A.; Filipchyk, A.; Klironomos, F.; Jara, C.A.C.; Fenske, P.; et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017, 357, eaam8526.
- Liu, J.; Li, P.; Wang, L.; Li, M.; Ge, Z.; Noordam, L.; Lieshout, R.; Verstegen, M.M.; Ma, B.; Su, J.; et al. Cancer-associated fibroblasts provide a stromal niche for liver cancer organoids that confers trophic effects and therapy resistance. Cell. Mol. Gastroenterol. Hepatol. 2020.
