1. Expression of SAPAPs in the CNS
During postnatal development, the different SAPAP members are expressed differently. The temporal patterns of
Sapap1 and
Sapap2 expression in individual brain regions do not change prominently after birth [
19]. For example, the expression of
Sapap1 mRNAs in the cortex seems to slightly peak around postnatal day 90. However, the spatiotemporal expression pattern of
Sapap3 during early postnatal development is noticeable, with peak expression observed around 2–3 weeks after birth in the cortex, striatum, and thalamus [
19,
71]. Although high expression levels are seen in the cortex within the first 3 weeks after birth, the distribution of
Sapap4 mRNA appears to slightly decline afterwards [
19]. Additionally, recent studies have indicated that both
Sapap1 and
Sapap4 are expressed in progenitor cells—the latter notably strongly expressed in the cortical progenitors and migrating neurons—suggesting the important roles of SAPAPs during early corticogenesis [
34,
72].
In the adult rodent brain, the
Sapap mRNAs and proteins display overlapping yet distinct distribution characteristics. All four
Sapap mRNAs are expressed abundantly in the cerebral cortex, hippocampus, and olfactory bulb, but at low or undetectable levels in the hypothalamus and substantia nigra [
18,
19].
Sapap1 mRNA is expressed abundantly in the cortex, hippocampus, amygdala, cerebellum, and olfactory bulb, moderately in the thalamus, and at low levels in the striatum [
13,
18]. Notably,
Sapap1 mRNA is only expressed in a small subset of cells in the striatum and displays heterogeneous expression patterns in different subregions of the thalamus [
18], but it is uniformly distributed throughout the whole cerebral cortex.
Distinct from the other SAPAP family members,
Sapap2 mRNAs are expressed in a limited pattern—mainly in the forebrain, with relatively low expression levels.
Sapap2 displays its highest expression levels in the hippocampus, with moderate levels in the striatum and almost undetectable levels in the cerebellum, thalamus, and amygdala [
13,
18]. In the cortex,
Sapap2 mRNAs show relative strong expression in layers 2 and 3 [
18].
The most abundant
Sapap detected in the striatum and various thalamic nuclei is
Sapap3 [
18,
19]. Moreover,
Sapap3 mRNAs are also highly expressed in layers 1–3 of the cortex, but only moderately expressed in the amygdala [
13,
18]. An intriguing aspect of the subcellular distribution of
Sapap3 is its dendritic localization. It has been found that several mRNAs are localized in the dendrites [
73,
74], and an extensive body of literature directly linked synaptic plasticity with local protein synthesis and degradation in dendrites [
75,
76,
77] (reviewed in [
73,
78,
79,
80]). In the hippocampus,
Sapap3 is the only family member that is highly localized in dendrites [
18,
19], which possibly allows rapid turnover of proteins and thus provides a mechanism for quick, activity-dependent changes in synaptic protein complex composition [
73,
80,
81], suggesting that SAPAP3 may play a unique role in synaptic plasticity.
Sapap4 is highly expressed in many regions of the brain, such as the hippocampus, cortex, amygdala, and cerebellum [
13,
18,
48]. In the hippocampus, the expression of
Sapap4 mRNA and proteins is relatively high in CA1 and CA3, but low in the dentate gyrus. Additionally,
Sapap4 mRNA shows high expression in layer 5 of the cortex. In addition to these areas,
Sapap4 is also strongly expressed in the adult rodent thalamus [
17,
18] and striatum [
13,
82]—second only to
Sapap3. Of particular interest,
Sapap4 mRNA is strongly expressed in the locus coeruleus [
18] as well as the ventricular zone and migrating neurons [
34].
In general, the SAPAP proteins are predominantly localized at glutamatergic and cholinergic synapses, and not at GABAergic or glycinergic synapses [
18], suggesting that SAPAPs may be a general “core” postsynaptic component of excitatory synapses, but not of inhibitory synapses. Moreover, subsequent studies have found that SAPAP3 and SAPAP4 may tend to present at corticostriatal and thalamostriatal excitatory synapses, respectively [
13,
82]. These findings of molecular heterogeneity of SAPAPs at different synapses imply that SAPAP family members may be localized at synapses in a circuit-selective manner, and that the molecular specificity in SAPAP proteins attributes a unique and specific function to the SAPAP family members. A systematic analysis of the SAPAPs according to the subregions and developmental stages of the brain remains to be determined and would be important for understanding the functions of SAPAPs in the brain.
2. SAPAPs’ Expression, FMRP, and Neuropsychiatric Disorders
Substantial studies have shown the genetic and epigenetic dysregulation of
SAPAP genes in patients afflicted with psychiatric disorders or related animal models [
21,
22,
23,
112,
113,
114,
115,
116]. Fragile X mental retardation protein (FMRP) is considered to be a translational repressor that plays a key role in regulating local mRNA translation in response to synaptic signaling. In
Fmr1-knockout (KO) mice—a model for fragile X syndrome—the protein levels of several FMRP targets are increased in PSD fractions either from the neocortex or hippocampus, including
Sapap1–3,
Shank1,
Shank3, and various glutamate receptor subunits [
117]. Conversely,
Sapap3 is decreased in both the OFC and medial prefrontal cortex (mPFC) of
Fmr1-KO mice [
118], which may contribute to the deficits in cognitive flexibility found in fragile X syndrome. Additionally, aggressive experience also increases the expression of
Sapap3, coupled with a decrease in FMRP phosphorylation that is dependent on the activation of mGluR5 [
119]. The postsynaptic targets of FMRP also include
Sapap4, mGluR5, and many other scaffold proteins that comprise the glutamate receptor interactomes, such as PSD-95, Homer1, and
Neuroligins 1–3 [
120,
121,
122]. The mRNA for
Sapap4 is significantly increased by the activation of mGluRs in WT but not
Fmr1-KO neurons [
123]. Diminished mGluR-induced dendritic localization of
Sapap4 mRNA is found in the hippocampal neurons of
Fmr1-KO mice [
123], indicating a surprising coordinate regulation between
Sapap4 and mGluRs that may be altered in fragile X syndrome. In accordance with these findings, increased mGluR5-signaling-dependent AMPAR endocytosis or altered mGluR5–Homer scaffolds are observed in OCD models with
Sapap3-mutant mice, fragile X syndrome models with
Fmr1-KO mice, and autism models with
Shank3-mutant mice, which all include repetitive behaviors, among other symptoms [
95,
101,
124,
125], suggesting a potential shared genetic mechanism of different neuropsychiatric disorders (
Figure 1).
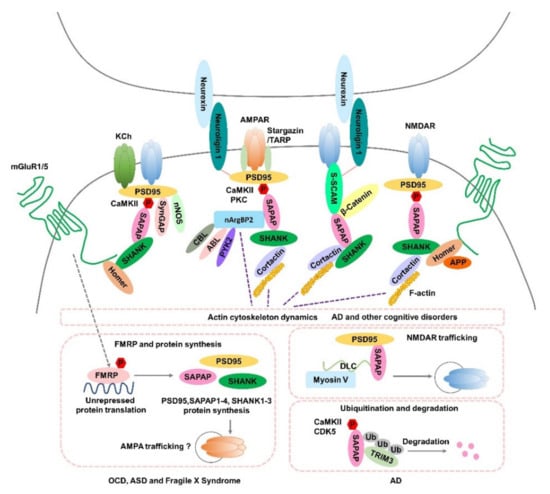
Figure 1. Potential roles of SAPAPs in synaptic function and the pathogenesis of neuropsychiatric disorders. Hypothesized mechanisms of SAPAPs for the molecular signaling underlying OCD, ASD, Fragile X Syndrome, AD and other cognitive disorders are summarized in the figure. AMPAR, α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor; NMDAR, N-methyl-D-aspartic acid receptor; KCh, Shaker-type K+ channels; SAPAP, SAP90/PSD-95-associated protein; mGluR1/5, Group I metabotropic glutamate receptors; CaMKII, Calcium/calmodulin-dependent protein kinase II; PKC, Protein kinase C; SynGAP, Synaptic GTPase Activating Protein; nNOS, Neural Nitric Oxide Synthase; S-SCAM, synaptic scaffolding molecule; nArgBP2, Neural Abelson-related gene-binding protein 2; CBL, E3 ubiquitin ligase casitas B-lineage lymphoma; ABL, tyrosine kinase; PYK2, proline-rich tyrosine kinase 2; APP, amyloid precursor protein; P, phosphorylation; FMRP, fragile X mental retardation protein; TRIM3, Tripartite motif-containing 3; Ub, ubiquitin; DLC, dynein light-chain; CDK5, cyclin-dependent kinase 5; TRAP, transmembrane AMPAR regulatory protein.
Altered expression levels of
SAPAPs are also observed in different neuropsychiatric disorders (
Table 1).
Sapap1 expression in the NAc is significantly increased in both animal models of phencyclidine-induced schizophrenia and unmedicated patients [
115].
Sapap2 mRNA expression, which is significantly associated with its DNA methylation status, was associated with coping with stress in an animal model of post-traumatic stress disorder (PSTD) [
112]. Similarly, the effects of
Sapap2 methylation on memory ability in AD patients are also thought to be mediated by changes in
SAPAP2 expression in the dorsal lateral prefrontal cortex (dlPFC) [
22]. Increases in
Sapap3 levels have been observed in both epilepsy patients and murine models [
116].
Sapap4 has been indicated to be involved in psychostimulants’ actions and bipolar disorder (BD). In taste-aversion-resistant rats after cocaine exposure, the mRNA levels of
Sapap4 in the amygdala were strikingly lower than those in taste-aversion-prone rats [
126]. Remarkably, sensitivity to cocaine and amphetamine was also changed in
Sapap4-deficient mice [
17,
35]. Interestingly, in human neural progenitor cells derived from BD patients,
Sapap4 has been identified as a target gene of miR-1908-5p-a BD-associated epigenetic regulator [
127]—suggesting that epigenetic dysregulation of
SAPAP4 may be involved in the pathogenesis of BD.
Epigenetic dysregulation of the expression of
SAPAP genes, mainly mediated by changes in DNA methylation and histone acetylation, may confer risks of various neuropsychiatric disorders (
Table 2). Epigenetic modifications of the
Sapap2 gene have been implicated in PTSD [
112], alcohol and cannabis use [
21,
114], schizophrenia [
113], and AD [
22]. One study revealed that histone deacetylases and methyl-CpG-binding protein 2 (
MeCP2) inhibit repetitive behaviors through regulation of the
Sapap3 gene [
128]. Additionally, recent studies have also reported an association between altered methylation of
SAPAP1 and stress responses to violence [
129] and a link between epigenetic dysregulation of
SAPAP4 and early-onset cerebellar ataxia [
23,
130].
3. Human Molecular Genetic Analysis of Neuropsychiatric Disorders Linked to SAPAP
3.1. SAPAP1 and Neuropsychiatric Disorders
Several human molecular genetic studies have emerged to show that
SAPAP1 is related to several neuropsychiatric disorders, including schizophrenia, OCD, AD, ADHD, ASD, mood disorders, and cortical malformation [
24,
25,
26,
27,
28,
131,
132,
133,
134] (
Table 3). The
SAPAP1 gene is located on chromosome 18p11, which has been suggested to be a susceptibility region for schizophrenia and/or BD [
135,
136]. Although several SNPs found in
SAPAP1 are not associated with schizophrenia [
131,
132], a de novo CNV mutation of
SAPAP1 has been identified in patients with schizophrenia [
24]. The first genome-wide association study (GWAS) with 1465 OCD cases revealed that the two SNPs with the lowest
p-values were located within
SAPAP1, suggesting a subthreshold statistical significance for
SAPAP1 [
25]. Furthermore, the fact that two rare CNVs (a 62 kb duplication for
SAPAP1 and a 16 kb deletion for
SAPAP2) overlapping the FMRP targets
SAPAP1 and
SAPAP2 were uncovered in another OCD study is of particular interest [
26]. Two
SAPAP1 SNPs associated with cognitive flexibility have also been identified in ADHD children [
27]. In addition,
SAPAP1 is also implicated in ASD [
133] and recurrent major depressive disorder (MDD) [
134]. Interestingly, the expression of
SAPAP1, which is significantly downregulated in the hippocampus and cortex of AD patients, is strongly associated with early identified AD variants (i.e., noncoding SNP rs8093731 in
Desmoglein-2/
DSG2 that acts as an expression quantitative trait loci for
SAPAP1) [
28] and
SAPAP1 is identified as a potential candidate gene involved in AD.
3.2. SAPAP2 and Neuropsychiatric Disorders
Numerous studies have implicated
SAPAP2 as being the strongest candidate gene for neurodevelopmental or behavioral phenotypes and closely associated with various neuropsychiatric disorders [
2,
20,
22,
26,
29,
30,
137,
138,
139,
140,
141,
142,
143,
144]. Strikingly, many genetic variants—including SNPs, single-nucleotide variants (SNVs), and de novo common and/or rare CNVs—in
SAPAP2 have been found to be associated with neurodevelopmental disorders such as ASD and ADHD [
20,
29,
30,
137,
138,
139,
140,
141,
142,
145], cognitive disorders such as developmental delay (DD)/ID and AD [
22,
142,
143,
145], and other disorders, such as schizophrenia [
2,
29,
139,
146,
147,
148] and OCD [
26,
144].
Although
SAPAP2 has been identified as a promising candidate gene of ASD, the potential role of
SAPAP2 in ASD is still unclear. A study using a murine model demonstrated that the deletion of
SAPAP2 leads to increased social interaction and aggressive behavior, along with impaired initial reverse learning and synaptic function [
14], indicating that
SAPAP2 somehow modulates cognition. Correspondingly, a recent cross-species study not only revealed the role of
SAPAP2 in age-related working memory decline, but also established an association between
SAPAP2 and AD phenotypes at multiple levels of analysis [
22], highlighting the possible importance of epigenetic dysregulation of
SAPAP2 expression in the pathophysiology of AD. As noted in the above studies, CDK5—one of the key kinases in AD—is involved in triggering amyloid-beta (Aβ)-induced ubiquitination and degradation of SAPAPs [
70], while amyloid precursor protein (APP) is one of binding partners of Homer, which is indirectly linked to SAPAPs via SHANKs [
7]. Gene overexpression can arise from de novo CNV duplication, and several autism risk genes in duplicated loci—such as
SAPAP2, SHANK3, and
Neuroligins 1 and 3—are all FMRP targets [
29,
117,
121,
138,
140], suggesting a promising link between dysfunction of FMRP in regulating the expression of synaptic proteins and the development of ASD.
3.3. SAPAP3 and Neuropsychiatric Disorders
After the initial report of the features of
Sapap3-mutant mice for OCD-like repetitive self-grooming behaviors [
13], a higher frequency of rare nonsynonymous coding variants located in
SAPAP3 was found in OCD patients, providing the first clinical evidence and tentatively supporting the link between
SAPAP3 and OCD [
152]. Several follow-up human genetic studies and other secondary analyses of GWAS have also noted that
SAPAP3 is a promising functional candidate gene for OCD and obsessive–compulsive spectrum disorders, such as trichotillomania (TTM) and Tourette syndrome (TS) [
31,
32,
33,
152], although some of the mutations identified in the above studies are more commonly found in association with grooming disorders and TTM than with OCD. Convergent evidence from functional imaging and neuropsychological studies has linked OCD symptoms to dysfunction of striatum-based circuitry, especially highlighting cortico-striato-thalamo-cortical circuitry [
5,
31,
154]. Owing to their robust behavioral similarities and sophisticated neural circuitry shared with OCD and related disorders [
13,
82],
Sapap3-mutant mice are now considered to be a well-established animal model for OCD and related disorders. Interestingly, a CNV within the
SAPAP3 gene was also identified in a study of about 200 individuals with ASD [
30]—a spectrum of disorders that involve prominent compulsive-like repetitive behaviors overlapped with the behavioral phenotypes of OCD.
3.4. SAPAP4 and Neuropsychiatric Disorders
In addition to the aforementioned possible roles in the actions of psychostimulants, BD, and fragile X syndrome,
SAPAP4 is also associated with several other neuropsychiatric disorders, including ASD, early-onset cerebellar ataxia, and subcortical heterotopias [
23,
34,
153]. An SNP located at the 20q11.21–q13.12 locus that encompasses the
SAPAP4 gene has also been linked with a potential role in the development of ASD [
153].
SAPAP4 is also implicated in early-onset cerebellar ataxia, according to an epigenetic study in which disruption of the
SAPAP4 gene by the chromosome translocation t (8;20) caused monoallelic DNA hypermethylation of the truncated
SAPAP4 promoter CpG island 820 bp downstream of the first untranslated exon of
SAPAP4 isoform a and perturbed expression of the
SAPAP4 gene [
23]. A recent study has identified a de novo frameshift variation and a missense variant in
SAPAP4 from two families with different types of subcortical heterotopias—which are cortical malformations associated with ID and epilepsy [
34]—pinpointing a novel role of
SAPAP4 in early cortical development. Although there is only an indirect link between
SAPAP4 and mood disorders such as BD [
127], 22q13 duplications spanning
SHANK3 (a known interaction scaffold protein of SAPAP4) have been found in patients diagnosed with ADHD and BD, respectively [
155], suggesting a possible role of
SAPAP4 in hyperkinetic disorders and mood disorders.
This entry is adapted from the peer-reviewed paper 10.3390/cells11233815