Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Plant Sciences
CRISPR/Cas techniques were mostly used against viral infection and for fungal and bacterial disease resistance. The CRISPR/Cas system has been used to develop resistance to many pathogen species.
- CRISPR/Cas9
- CRISPR/Cas12
- CRISPR/Cas13
- base editing
1. Introduction
Plants produce food, fuel, and feed, which are essential in daily human and animal life for nourishment and growth. In the process of plant growth, they will be affected by a variety of biological stresses (bacteria, viruses, fungi, and insects) and abiotic stresses [1][2][3][4][5]. Due to continuous global climate change and anthropogenic activity, the impact of abiotic stresses on crop growth and development is becoming more serious. Abiotic stresses, including drought, salinity, waterlogging, heat/cold, and heavy metals, significantly reduce agricultural production worldwide. Therefore, the ability to breed new species that are tolerant to various stresses in order to reduce yield loss will be a sustainable way to overcome these obstacles and meet the growing needs of human beings. Different types of biotic stresses involve a complex interplay among pathogens and host plants based on the susceptibility/resistance of crop plants to any disease. The latest advances in molecular tools have provided insights into a wide array of pathogen infection mechanisms and their interactions with specific crop plants. The insertions/deletions (Indels) mutations by artificial or natural phenomena might be involved in altering these interactions and hinder the pathways involved in the mode of infection [6][7].
Traditional crop breeding such as crossbreeding is an effective method that has been widely used to modify various plant species. Crop productivity and varieties can be increased effectively through crop breeding programs. In modern agriculture, the key methodologies used for breeding purposes are transgenic breeding, mutation-breeding, and GE-mediated breeding for crop improvement [8]. Cross-breeding and genetic recombination require years to introduce desirable alleles and increase variability [8]. Transgenic breeding is easy and well-known, improving crop traits by the exogenous transformation of genes into economically important elite varieties greatly shortens the breeding time. Still, this method inserts specific genes into the genome at random locations through plant transformation, which results in varieties containing foreign DNA. Compared to crossbreeding, mutation-breeding, and traditional transgenic breeding, GE-mediated crop breeding is fast, efficient, and accurate (Figure 1). GE improves a targeted trait in a very fast and short time and exactly revising the target gene or regulatory sequence or changing the DNA and/or RNA bases in elite varieties. The current GE technique includes meganuclease (MegN), zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) (CRISPR/Cas9) [7][9][10][11]. In 2013, genetic modifications through the CRISPR/Cas9 method were developed in plants and revolutionized the field by eliminating the barriers to targeted GE. The CRISPR system has been used in wheat, rice, tobacco, and Arabidopsis thaliana [12][13][14][15][16][17][18][19][20][21][22][23][24][25]. Till now, GE has been practiced in more than 50 plant species, and it will revolutionize plant breeding [26][27][28][29][30][31][32][33][34][35][36][37][38][39][40].
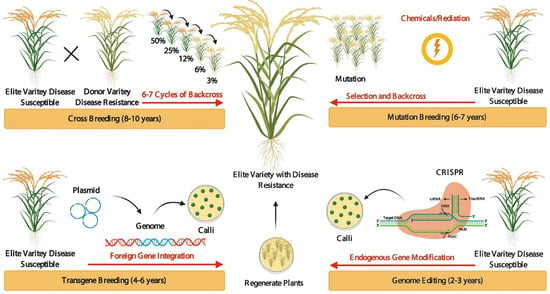
Figure 1. Evolution of crop breeding techniques. Crossbreeding takes a great deal of time (8–10 years) to improve desirable characters/traits (in a particular species for disease tolerance or resistance) through crossing an elite variety line with a donor variety line and selecting the new outstanding offspring with desirable characters/traits. To introduce new progeny with desirable traits from the donor variety line to the elite variety line, the selected offspring must be backcrossed to the elite variety line for several years to remove undesirable related traits. In mutation breeding, mutations are used to improve traits in the time (6–7 years) of the genome through chemical treatment or physical irradiation to create novel genetic variations. Transgenic breeding is easy and well-known, improving crop traits within (4–6 years) by the exogenous transformation of genes into economically important elite varieties. Genome editing: improving a targeted trait in a very fast and short time (2–3 years) and exactly revising the target gene or regulatory sequence or changing the DNA and/or RNA bases in elite varieties.
Based on the composition of the CRISPR locus, this system has been divided into two classes: Class 1 requires multiple effector proteins with subtypes I, III, and IV, while class 2 requires only a single effector protein with subtypes II, V, and VI. The mode-of-action of GE by site-directed nucleases (SDNs) is that once present in a cell by insertion/expression and or transfection, the SDN is capable of cutting the genome at a targeted site. The cellular DNA-repair mechanisms fix the cut sites either by the non-homologous end joining (NHEJ) or by homology-directed repair (HDR). As NHEJ can be an error-prone process, indels can appear at the respective genomic site, leading to a loss-of-function edited gene sequence due to frameshift mutations. GE by using SDNs, can be categorized into three types: SDN-1 introduces small insertions or deletions which carry no additional or recombinant DNA. SDN-2 introduces short insertions or editing of a few base pairs by an external DNA-template sequence. The SDN-3, using a similar method to SDN-2, can be considered transgenic due to the insertion of large DNA pieces [41][42]. Since its introduction, in recent years, constant improvements have been made to make CRISPR systems easier and more suitable for different constraints, such as CRISPR/Cas9 [12][13][43][44], CRISPR/Cas12a [45][46][47][48][49], CRISPR/Cas12b [50], CRISPR/Cas13 [51][52], base editing tools [53][54][55][56][57][58][59], and CRISPR transcriptional activation (CRISPRa) [60][61][62][63][64] (Figure 2). A new form of GE technology, known as Prime Editing (PE) has recently been developed which is capable of achieving various forms of editing, for example, some base-to-base transfer, such as all transformations (C→T, G→A, A→G, and T→C) and transversion mutations (C→A, C→G, G→C, G→T, A→C, A→T, T→A, and T→G), as well as small indels without double-stranded breaks in the DNA. Since PE has enough versatility to accomplish specific forms of editing in the genome, it has great potential to grow superior crops for different purposes, including production, avoiding various biotic and abiotic stresses, and enhancing the quality of plant products [57][58][65][66][67][68][69][70][71][72][73].
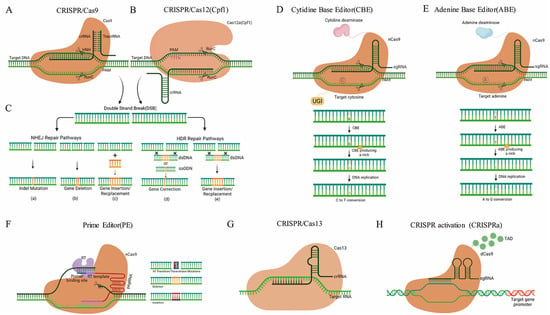
Figure 2. The methodology of major CRISPR/Cas systems. (A) CRISPR/Cas9 induces double-stranded breaks (DSBs) in DNA strands. (B) CRISPR/Cas12a cleaves the target DNA and introduces DSBs. (C) CRISPR/Cas methods can achieve different research goals: (a–c) are results of non-homologous end-joining NHEJ, and (d,e) are results of the homology-directed repair HDR repair pathways using a donor DNA template. (D–F) Base editing tools mainly include Cytidine Base Editor (CBE), Adenine Base Editor (ABE), and Prime Editor (PE). (D) CBE converts C-G base pairs to T-A base pairs at the target site. (E) ABE converts A-T base pairs to G-C base pairs at the target site. (F) PE is a new base editing system, which enables precise sequence substitution, insertion, and deletion. PE mainly consists of a Cas9 nickase (nCas9), an engineered reverse transcriptase (RT), and pegRNA. PegRNA includes PBS (Primer Binding Site) sequence and RT Template. (G) CRISPR/Cas13 consists of a Cas13, a crRNA, and a target RNA. Cas13:crRNA complexes bind target RNA and cleave the target RNA. (H) CRISPR transcriptional activation (CRISPRa) consists of a nuclease-deficient Cas9 (dCas9) and transcription activation domain (TAD). CRISPRa activates the transcription of single or multiple target genes.
CRISPR/Cas method has become the most popular among editing technologies and, thus far, has revealed the greatest potential to overcome the developing challenges (such as yield and biotic and abiotic stresses) of agriculture [9][74][75][76]. For example, mutations conferring resistance to various diseases in lettuce also exist [77]. Resistance against powdery mildew has been successfully acquired in barley by creating mutants at the mildew resistance locus o (MLO) [78]. The mutation at MLO is remarkable because it provides extraordinary, stable, and precise resistance for two decades against mildew without breakage of alleles; this long-lasting resistance is because of gene knockout [79][80].
2. CRISPR/Cas Technique for Disease Resistance
Biotic stresses, such as bacterial, viral, and fungal diseases, as well as herbivores, damage plant products every year, affecting 11% to 30% of worldwide agriculture production [81]. Plant defense against pathogens can reduce the effects of disease on plant growth and productivity, which is highly relevant to the lack of food availability in the world with the increasing population. Improvements in new methods or GE techniques have improved the new resistant crops, reducing yield losses due to plant defense. Until now, CRISPR/Cas techniques were mostly used against viral infection and for fungal and bacterial disease resistance (Figure 3). The CRISPR/Cas system has been used to develop resistance to many pathogen species [26][82].

Figure 3. Future applications of CRISPR/Cas in plants against the biotic and abiotic stress. CRISPR/Cas represents the future of genome editing technology and the potential use of the CRISPR/Cas system in various disciplines under biotic and abiotic stresses of agriculture. With the maturity of genome editing (GE) technology and the development of new GE tools, the application of CRISPR/Cas is becoming more and more extensive. CRISPR/Cas can now achieve gene knockout, knock-in, and knock-up in plants, replacing a single base to cause amino acid changes, etc. Therefore, CRISPR/Cas can be used to modify key genes of biotic and abiotic stresses, improving crop growth and development and coping with various environmental stresses to create more germplasm resources that meet human needs.
2.1. CRISPR/Cas-Mediated Fungal Resistance in Plants
Many fungal pathogens cause lethal diseases in crop plants, such as rust, mildew, rot, and smut, which not only damage yield yearly in the biosphere but also damage the quality of the product. CRISPR/Cas has improved mycological resistance in various crop species based on the available information of the genomic mechanisms involved in crop-pathogen interactions. Defined candidate genes and gene products have provided the potential to increase plant defense against fungi [83][84]. In three crop varieties, RNA-guided Cas9 endonuclease was used to target MLO loci, such as tomato (Solanum lycopersicum), grapevine (Vitis vinifera), and wheat [85][86][87][88][89], and transgene-free plants have been generated [90]. An MLO encoded protein is localized in the cell membrane and contains seven transmembrane domains, which universally exist in all dicots and monocots [91]. Plants carrying loss-of-function alleles (mlo) of the MLO, such as A. thaliana, tomato, and barley, confer durable resistance against powdery mildew [92][93][94]. Using precision GE to target the MLO-B1 locus of the wheat genome to generate a 304K deletion Tamlo-R32 mutant maintains wheat growth and yield while providing robust powdery mildew resistance [79]. Out of three MLO home alleles, one (TaMLO-A1) has been mutated by CRISPR/Cas9 in triticum aestivum and displayed resilient resistance against Blumeria graminis f. sp. tritici infection [85]. The CRISPR/Cas-mediated transgene-free and self-pollinated tomato variety, which was developed by deleting the 48 bp fragment in the SlMlo1 gene (out of 16 important SlMlo genes), offers resistance against powdery mildew Oidium neolycopersici [87].
In grapevine, loss of VvMLO7 function by RNAi reduced sensitivity against powdery mildew Erysiphe necator [95]. In parallel, the knockout of VvMLO7 and VvMLO3 using CRISPR/Cas9 enhanced resistance to powdery mildew in grapevine [88][89]. In apple (Malus Domestica) protoplasts, RNP-based technology has been successfully used to edit three (DIPM-1, DIPM-2, and DIPM-4) genes to create resistance against fire blight Erwinia amylovora [88]. CRISPR/Cas9 scheme was used to target the VvWRKY52 transcription factor with four guide RNAs. The results showed 21% biallelic mutations in regenerated plants, and these plants confer resistance to the fungus Botrytis cinerea compared with monoallelic mutant plants [96]. To accelerate the GE application in woody plants, another approach based on transient leaf transformation together with disease assays was first demonstrated by researchers in Theobroma cacao [97]. Pathogenesis-Related 3 (NPR3) gene (the immune system suppressor) was targeted in cacao leaves, transiently by CRISPR/Cas9 system, so the leaves showed enhanced resistance against Phytophthora tropicalis. GE of a fungicide resistant gene PcMuORP1 by CRISPR/Cas9 elucidates a novel selection marker for Phytophthora (a genus of oomycetes) species [98]. In rice, CRISPR/Cas9-mediated disruption of OsSEC3A and OsERF922 genes confer resistance against rice blast disease [99][100]. In addition, the pi21 gene in rice also induced durable resistance to rice blast [101]. Furthermore, resistance to Magnaporthe oryzae disease in rice was enhanced by generating the OsSEC3A mutants and showed a pleiotropic type of phenotype with an increase in salicylic acid (SA) concentration, and several genes were induced related to SA- and pathogenesis related genes [99]. To conclude, all these successful fungal disease resistance results determined the advantage, efficacy, and potential of the CRISPR/Cas-based editing system to enhance resistance in crop plants.
2.2. CRISPR/Cas-Mediated Viral Resistance in Plants
Plant viruses are among the most common pathogens and cause hazardous diseases in a variety of economically important crops. There are five main groups based on viral genomes characters: sense-single-stranded-RNA (ssRNA+), antisense-single stranded-RNA (asRNA-), single-stranded-DNA (ssDNA), double-stranded-DNA (dsDNA), and double-stranded-RNA (dsRNA) viruses [102]. A rolling-circle amplification system is required to replicate the virus genome through recombination-mediated duplication or by a dsDNA replicative form [103]. Their genome holds a mutual fragment of 220 bp, which is prearranged in one (A, monopartite) or two (A and B, bipartite) constituents [104]. The Geminiviridae are a large family (over 360 species) of ssDNA plant viruses that cause significant losses to agriculturally and economically important crop plants worldwide [103], such as Malvaceae, Solanaceae, Fabaceae, Euphorbiaceae, and Cucurbitaceae [105]. The commercial term for a large genus of geminiviruses is Begomoviruses. Begomoviruses mostly produce diseases in dicotyledonous plants, for example, Nicotiana tabacum and sweet potato (Ipomoea batatas), and these viruses are mostly transmitted via the whitefly or leafhopper [106][107]. CRISPR/Cas9 system was used in Nicotiana benthamiana and A. thaliana to target two different geminiviruses: Bean yellow dwarf virus (BeYDV) and Beet severe curly top virus (BSCTV), respectively [108][109]. Recently, CRISPR/Cas9 techniques have also been applied to attain resistance against Begomoviruses [110]. In the (BSCTV) genome, 43 candidates were selected to target their coding and non-coding regions using CRISPR/Cas9 [109]. In inoculated leaves, virus accumulation was significantly reduced in all CRISPR/Cas9 constructs at variable levels. However, the highest resistance was observed in A. thaliana and N. tabacum to virus infection displaying the maximum expression level of sgRNAs and Cas9. Similar results have been detected by employing 11 sgRNAs in N. benthamiana, targeting the non-nucleotide sequence, Rep-binding sites, Rep motifs, and the hairpin of BeYDV [108], and decreased up to 87% load of the targeted viral. A tobacco rattle virus (TRV) vector was used to deliver the sgRNA molecules to the N. benthamiana, stably overexpressing the Cas9 endonuclease to target the Tomato yellow leaf curl virus (TYLCV) genome [110]. In that study, the CRISPR/Cas approach was effectively implemented to cleave and target the virus genome during duplication to confer resistance against TYLCV [86][110][111].
By using specific sgRNAs, several genome loci of TYLCV (non-coding and coding sequences) were targeted in their intergenic region (IR), the RCRII motif replication protein (Rep), and the viral capsid protein (CP). Targeting the IR stem-loop invariant structure showed the lowest viral accumulation and replication [110]. A similar CRISPR/Cas9 system was established to target the geminiviruses monopartite beet curly top virus (BCTV), and bipartite Merremia mosaic virus (MerMV), which possess a similar IR stem-loop sequence. CRISPR/Cas9 system-edited BCTV and MerMV viruses displayed tempered symptoms, indicating that combined resistance against various viral strains can be achieved by a single sgRNA specific for the conserved region of the pathogen.
The traditional SpCas9 system recognizes only dsDNA, so the defense against RNA-based viruses is difficult to attain. Nevertheless, the characterization and search for associated nucleases have steered to the discovery of LwaCas13a from (Leptotrichia wadei) and FnCas9 from (Francisella novicida), which have the ability to bind and cut the RNA [112]. FnCas9 was reported to demonstrate resistance against RNA viruses [113]. The sgRNAs designed to target the RNA of cucumber mosaic virus (CMV) and tobacco mosaic virus (TMV) in N. benthamiana and A. thaliana transgenic plants showed a significant reduction in TMV and CMV by 40–80% compared to wild-type (WT) plants [113]. It demonstrated that FnCas9-mediated application could be deliberated as a CRISPR interference (CRISPRi) apparatus, similar to the mitigation of gene expression by catalytically inactive proteins of SpCas9 [114]. A similar study was carried out with Cas13a for manipulating the RNA genome of turnip mosaic virus (TuMV) using RNA-guided ribonuclease [115]. The minimum spread and replication of TuMV was observed in tobacco leaves by using the most proficient virus interference, detected with CRISPR RNA excision of GFP2 and HC-Pro genes.
Furthermore, the pre-CRISPR RNA was processed by Cas13 (due to its innate ability) into functional CRISPR RNA to target many viral mRNAs simultaneously. This may provide an alternative system to improve its efficiency distinctly [115][116][117]. A second strategy is to achieve viral resistance by editing the specific plant genes that are responsible for virus resistance traits [118][119][120]. RNA viruses need plant host factors to preserve their normal life cycle, containing the eukaryotic translation initiation factors eIF4E, eIF4G, and eIF(iso)4E [121]. Host susceptibility gene eIF4E was targeted at two different sites to create resistance against plant potyviruses by CRISPR/Cas9 [118][122][123]. A similar approach in A. thaliana plants induced site-specific mutations at eIF(iso)4E locus and conferred complete resistance to single-stranded RNA potyvirus -TuMV by 1 bp deletions and 1 bp insertions without any off-target modification [119]. Recently, resistance to rice tungro spherical virus (RTSV) was developed by the mutagenesis in eIF4G alleles [120]. In addition, no negative effects on the growth of mutant plants were observed in studies by Macovei et al. and Pyott et al., although additional research should be conducted to verify and test the durability and efficacy of recessive resistance edited plants [119][120].
2.3. CRISPR/Cas-Mediated Bacterial Resistance in Plants
Many pathogenetic bacteria cause diseases in crops, and the crops show several types of symptoms [124]. Compared to fungal and viral resistance, few studies have been reported about the utilization of CRISPR/Cas against bacterial diseases in crop plant species. The Xanthomonas oryzae pv. Oryzae causes host gene expression to induce susceptibility by utilizing the type III transcription-activator-like effectors (TALEs) system. The X. oryzae pv. oryzae effector protein PthXo2 targets the host sucrose transporter gene OsSWEET13 and is recognized as a sensitive gene for pathogen progression. Disease susceptibility was conferred by transferring the indica rice IR24 OsSWEET13 allele to japonica rice Kitaake, while CRISPR/Cas9-mediated mutations in the allele offered resistance against bacterial blight [125]. Recently, a mutation in the promoter of three rice genes confers broad-spectrum resistance against bacterial blight in rice [126]. CRISPR/Cas9 was used to edit the promoter of the Xa13, a pluripotent gene for recessive resistance to bacterial blight in rice to obtain the highly resistant rice that does not affect agronomic traits [127]. Downy mildew resistance 6 (DMR6) is a well-known negative regulator of plant defense. In tomato, DMR6 ortholog SlDMR6-1 was reported to be up-expressed during Pseudomonas syringae pv. tomato pathogen progression and Phytophthora capsici infection [128]. By targeting the SlDMR6-1 (exon-3), the mutated plants conferred wide-spectrum resistance against P. capsici, Xanthomonas gardneri, P. syringae, and X. perforans [128][129][130]. The tomato bacterial speck disease (causal agent Pseudomonas syringae) causes stomatal opening using coronatine (COR) to facilitate bacterial progression. This stomatal response in A. thaliana relies on AtJAZ2 (Jasmonate ZIM-domain-2), a COR co-receptor. The JAZ2 does not have the C-terminal Jas domain (JAZ2Δjas) that avoids stomatal opening using COR [131]. The homologous gene of AtJAZ2 in tomato is SlJAZ2 [132]. Resistance against the model pathogen Pseudomonas syringae pv. tomato DC3000 (Pto) DC3000 was developed by targeting the dominant JAZ2 repressor- SlJAZ2Δjas by using CRISPR/Cas9 technology that prohibited stomatal opening. Improving and refining the CRISPR/Cas9 and CRISPR/Cas12a systems provide a new opportunity to edit perennial crops species such as citrus to introduce resistance against citrus greening disease [133].
After producing successful bacterial disease-resistant tomato and A. thaliana, the CRISPR/Cas9 system recently effectively produced citrus bacterial canker (CBC) (causal agent Xanthomonas citri subsp. citri (X. citri) resistant citrus plants. The X. citri is the most widespread disease in commercially cultivated citrus [134]. CBC resistance was firstly reported in Duncan grapefruit by altering the PthA4 effector binding elements in the promoter of the Lateral Organ Boundaries 1 (CsLOB1) gene [135]. A significant decline in Xcc symptoms was detected in the mutated lines with no additional phenotypic alterations confirming the link between CBC disease susceptibility and CsLOB1 promoter activity Citrus (Citrus sinensis) (Osbeck) Wanjincheng orange [136]. In Wanjincheng orange, editing of CsWRKY22 by CRISPR/Cas9 reduces susceptibility to X. citri [137]. CBC disease resistance was enhanced by deleting the EBEPthA4 sequence completely from both CsLOB1 alleles, and no additional changes were observed in plants with altered CsLOB1 promoter. Recently, the CRISPR/Cas9-FLP/FRT system has been successfully induced in apple cultivars to reduce fire blight susceptibility [138]. In conclusion, these fruitful results demonstrate that CRISPR/Cas has the potential to not only create bacterial resistance in annual and biennial crop species but also confer durable bacterial disease resistance in perennial crop plants.
This entry is adapted from the peer-reviewed paper 10.3390/cells11233928
References
- Borrelli, V.M.G.; Brambilla, V.; Rogowsky, P.; Marocco, A.; Lanubile, A. The Enhancement of Plant Disease Resistance Using CRISPR/Cas9 Technology. Front. Plant Sci. 2018, 9, 1245.
- Rodriguez-Leal, D.; Lemmon, Z.H.; Man, J.; Bartlett, M.E.; Lippman, Z.B. Engineering Quantitative Trait Variation for Crop Improvement by Genome Editing. Cell 2017, 171, 470–480.
- Shaibu, A.S.; Li, B.; Zhang, S.R.; Sun, J.M. Soybean cyst nematode-resistance: Gene identification and breeding strategies. Crop J. 2020, 8, 892–904.
- Landa, B.B.; Saponari, M.; Feitosa-Junior, O.R.; Giampetruzzi, A.; Vieira, F.J.D.; Mor, E.; Robatzek, S. Xylella fastidiosa’s relationships: The bacterium, the host plants and the plant microbiome. New Phytol. 2022, 234, 1598–1605.
- Dong, O.X.; Ronald, P.C. Genetic Engineering for Disease Resistance in Plants: Recent Progress and Future Perspectives. Plant Physiol. 2019, 180, 26–38.
- Dracatos, P.M.; Haghdoust, R.; Singh, D.; Park, R.F. Exploring and exploiting the boundaries of host specificity using the cereal rust and mildew models. New Phytol. 2018, 218, 453–462.
- Manghwar, H.; Lindsey, K.; Zhang, X.L.; Jin, S.X. CRISPR/Cas System: Recent Advances and Future Prospects for Genome Editing. Trends Plant Sci. 2019, 24, 1102–1125.
- Scheben, A.; Wolter, F.; Batley, J.; Puchta, H.; Edwards, D. Towards CRISPR/Cas crops—Bringing together genomics and genome editing. New Phytol. 2017, 216, 682–698.
- Manghwar, H.; Li, B.; Ding, X.; Hussain, A.; Lindsey, K.; Zhang, X.L.; Jin, S.X. CRISPR/Cas Systems in Genome Editing: Methodologies and Tools for sgRNA Design, Off-Target Evaluation, and Strategies to Mitigate Off-Target Effects. Adv. Sci. 2020, 7, 1902312.
- Zhang, D.; Hussain, A.; Manghwar, H.; Xie, K.; Xie, S.; Zhao, S.; Larkin, R.M.; Qing, P.; Jin, S.; Ding, F. Genome editing with the CRISPR-Cas system: An art, ethics and global regulatory perspective. Plant Biotechnol. J. 2020, 18, 1651–1669.
- Khalil, A.M. The genome editing revolution: Review. J. Genet. Eng. Biotechnol. 2020, 18, 68.
- Shan, Q.W.; Wang, Y.P.; Li, J.; Zhang, Y.; Chen, K.L.; Liang, Z.; Zhang, K.; Liu, J.X.; Xi, J.J.; Qiu, J.L.; et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686–688.
- Li, J.F.; Norville, J.E.; Aach, J.; McCormack, M.; Zhang, D.D.; Bush, J.; Church, G.M.; Sheen, J. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 2013, 31, 688–691.
- Xing, H.L.; Dong, L.; Wang, Z.P.; Zhang, H.Y.; Han, C.Y.; Liu, B.; Wang, X.C.; Chen, Q.J. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014, 14, 327.
- Hyun, Y.; Kim, J.; Cho, S.; Choi, Y.; Kim, J.S.; Coupland, G. Site-directed mutagenesis in Arabidopsis thaliana using dividing tissue-targeted RGEN of the CRISPR/Cas system to generate heritable null alleles. Planta 2015, 241, 271–284.
- Steinert, J.; Schiml, S.; Fauser, F.; Puchta, H. Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. Plant J. 2015, 84, 1295–1305.
- Jiang, W.Z.; Zhou, H.B.; Bi, H.H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013, 41, e188.
- Yin, K.Q.; Han, T.; Liu, G.; Chen, T.Y.; Wang, Y.; Yu, A.Y.L.; Liu, Y.L. A geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Sci. Rep. 2015, 5, 14926.
- Vazquez-Vilar, M.; Bernabe-Orts, J.M.; Fernandez-del-Carmen, A.; Ziarsolo, P.; Blanca, J.; Granell, A.; Orzaez, D. A modular toolbox for gRNA-Cas9 genome engineering in plants based on the GoldenBraid standard. Plant Methods 2016, 12, 10.
- Gao, J.P.; Wang, G.H.; Ma, S.Y.; Xie, X.D.; Wu, X.W.; Zhang, X.T.; Wu, Y.Q.; Zhao, P.; Xia, Q.Y. CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol. Biol. 2015, 87, 99–110.
- Mercx, S.; Tollet, J.; Magy, B.; Navarre, C.; Boutry, M. Gene Inactivation by CRISPR-Cas9 in Nicotiana tabacum BY-2 Suspension Cells. Front. Plant Sci. 2016, 7, 40.
- Miao, J.; Guo, D.S.; Zhang, J.Z.; Huang, Q.P.; Qin, G.J.; Zhang, X.; Wan, J.M.; Gu, H.Y.; Qu, L.J. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 2013, 23, 1233–1236.
- Xu, R.F.; Li, H.; Qin, R.Y.; Wang, L.; Li, L.; Wei, P.C.; Yang, J.B. Gene targeting using the Agrobacterium tumefaciens-mediated CRISPR-Cas system in rice. Rice 2014, 7, 5.
- Zhang, H.; Zhang, J.S.; Wei, P.L.; Zhang, B.T.; Gou, F.; Feng, Z.Y.; Mao, Y.F.; Yang, L.; Zhang, H.; Xu, N.F.; et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 2014, 12, 797–807.
- Du, H.Y.; Zeng, X.R.; Zhao, M.; Cui, X.P.; Wang, Q.; Yang, H.; Cheng, H.; Yu, D.Y. Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J. Biotechnol. 2016, 217, 90–97.
- Bisht, D.S.; Bhatia, V.; Bhattacharya, R. Improving plant-resistance to insect-pests and pathogens: The new opportunities through targeted genome editing. Semin. Cell Dev. Biol. 2019, 96, 65–76.
- Gao, C. Genome engineering for crop improvement and future agriculture. Cell 2021, 184, 1621–1635.
- Fauser, F.; Schiml, S.; Puchta, H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 2014, 79, 348–359.
- Schiml, S.; Fauser, F.; Puchta, H. The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J. 2014, 80, 1139–1150.
- Zhao, Y.P.; Zhang, C.S.; Liu, W.W.; Gao, W.; Liu, C.L.; Song, G.Y.; Li, W.X.; Mao, L.; Chen, B.J.; Xu, Y.B.; et al. An alternative strategy for targeted gene replacement in plants using a dual-sgRNA/Cas9 design. Sci. Rep. 2016, 6, 23890.
- Ron, M.; Kajala, K.; Pauluzzi, G.; Wang, D.X.; Reynoso, M.A.; Zumstein, K.; Garcha, J.; Winte, S.; Masson, H.; Inagaki, S.; et al. Hairy Root Transformation Using Agrobacterium rhizogenes as a Tool for Exploring Cell Type-Specific Gene Expression and Function Using Tomato as a Model. Plant Physiol. 2014, 166, 455–469.
- Ito, Y.; Nishizawa-Yokoi, A.; Endo, M.; Mikami, M.; Toki, S. CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem. Biophys. Res. Commun. 2015, 467, 76–82.
- Pan, C.T.; Ye, L.; Qin, L.; Liu, X.; He, Y.J.; Wang, J.; Chen, L.F.; Lu, G. CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci. Rep. 2016, 6, 24765.
- Shan, Q.W.; Wang, Y.P.; Li, J.; Gao, C.X. Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 2014, 9, 2395–2410.
- Liang, Z.; Zhang, K.; Chen, K.L.; Gao, C.X. Targeted Mutagenesis in Zea mays Using TALENs and the CRISPR/Cas System. J. Genet. Genom. 2014, 41, 63–68.
- Zhu, J.J.; Song, N.; Sun, S.L.; Yang, W.L.; Zhao, H.M.; Song, W.B.; Lai, J.S. Efficiency and Inheritance of Targeted Mutagenesis in Maize Using CRISPR-Cas9. J. Genet. Genom. 2016, 43, 25–36.
- Feng, C.; Yuan, J.; Wang, R.; Liu, Y.; Birchler, J.A.; Han, F.P. Efficient Targeted Genome Modification in Maize Using CRISPR/Cas9 System. J. Genet. Genom. 2016, 43, 37–43.
- Li, C.; Liu, C.; Qi, X.; Wu, Y.; Fei, X.; Mao, L.; Cheng, B.; Li, X.; Xie, C. RNA-guided Cas9 as an in vivo desired-target mutator in maize. Plant Biotechnol. J. 2017, 15, 1566–1576.
- Dong, L.; Li, L.; Liu, C.; Liu, C.; Geng, S.; Li, X.; Huang, C.; Mao, L.; Chen, S.; Xie, C. Genome Editing and Double-Fluorescence Proteins Enable Robust Maternal Haploid Induction and Identification in Maize. Mol. Plant 2018, 11, 1214–1217.
- Lawrenson, T.; Shorinola, O.; Stacey, N.; Li, C.D.; Ostergaard, L.; Patron, N.; Uauy, C.; Harwood, W. Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 2015, 16, 258.
- Gupta, S.; Kumar, A.; Patel, R.; Kumar, V. Genetically modified crop regulations: Scope and opportunity using the CRISPR-Cas9 genome editing approach. Mol. Biol. Rep. 2021, 48, 4851–4863.
- Menz, J.; Modrzejewski, D.; Hartung, F.; Wilhelm, R.; Sprink, T. Genome Edited Crops Touch the Market: A View on the Global Development and Regulatory Environment. Front. Plant Sci. 2020, 11, 586027.
- Lowder, L.G.; Zhang, D.W.; Baltes, N.J.; Paul, J.W.; Tang, X.; Zheng, X.L.; Voytas, D.F.; Hsieh, T.F.; Zhang, Y.; Qi, Y.P. A CRISPR/Cas9 Toolbox for Multiplexed Plant Genome Editing and Transcriptional Regulation. Plant Physiol. 2015, 169, 971–985.
- Wang, P.; Zhang, J.; Sun, L.; Ma, Y.; Xu, J.; Liang, S.; Deng, J.; Tan, J.; Zhang, Q.; Tu, L.; et al. High efficient multisites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system. Plant Biotechnol. J. 2018, 16, 137–150.
- Li, B.; Rui, H.; Li, Y.; Wang, Q.; Alariqi, M.; Qin, L.; Sun, L.; Ding, X.; Wang, F.; Zou, J.; et al. Robust CRISPR/Cpf1 (Cas12a)-mediated genome editing in allotetraploid cotton (Gossypium hirsutum). Plant Biotechnol. J. 2019, 17, 1862–1864.
- Li, B.; Liang, S.; Alariqi, M.; Wang, F.; Wang, G.; Wang, Q.; Xu, Z.; Yu, L.; Naeem Zafar, M.; Sun, L.; et al. The application of temperature sensitivity CRISPR/LbCpf1 (LbCas12a) mediated genome editing in allotetraploid cotton (G. hirsutum) and creation of nontransgenic, gossypol-free cotton. Plant Biotechnol. J. 2021, 19, 221–223.
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 2015, 163, 759–771.
- Shimatani, Z.; Kashojiya, S.; Takayama, M.; Terada, R.; Arazoe, T.; Ishii, H.; Teramura, H.; Yamamoto, T.; Komatsu, H.; Miura, K.; et al. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 441–443.
- Zhang, Y.; Ren, Q.; Tang, X.; Liu, S.; Malzahn, A.A.; Zhou, J.; Wang, J.; Yin, D.; Pan, C.; Yuan, M.; et al. Expanding the scope of plant genome engineering with Cas12a orthologs and highly multiplexable editing systems. Nat. Commun. 2021, 12, 1944.
- Wang, Q.; Alariqi, M.; Wang, F.; Li, B.; Ding, X.; Rui, H.; Li, Y.; Xu, Z.; Qin, L.; Sun, L.; et al. The application of a heat-inducible CRISPR/Cas12b (C2c1) genome editing system in tetraploid cotton (G. hirsutum) plants. Plant Biotechnol. J. 2020, 18, 2436–2443.
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.T.; Kellner, M.J.; Regev, A.; et al. RNA targeting with CRISPR-Cas13. Nature 2017, 550, 280–284.
- Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA editing with CRISPR-Cas13. Science 2017, 358, 1019–1027.
- Wang, M.; Xu, Z.; Gosavi, G.; Ren, B.; Cao, Y.; Kuang, Y.; Zhou, C.; Spetz, C.; Yan, F.; Zhou, X.; et al. Targeted base editing in rice with CRISPR/ScCas9 system. Plant Biotechnol. J. 2020, 18, 1645–1647.
- Qin, L.; Li, J.; Wang, Q.; Xu, Z.; Sun, L.; Alariqi, M.; Manghwar, H.; Wang, G.; Li, B.; Ding, X.; et al. High-efficient and precise base editing of C•G to T•A in the allotetraploid cotton (Gossypium hirsutum) genome using a modified CRISPR/Cas9 system. Plant Biotechnol. J. 2020, 18, 45–56.
- Wang, G.; Xu, Z.; Wang, F.; Huang, Y.; Xin, Y.; Liang, S.; Li, B.; Si, H.; Sun, L.; Wang, Q.; et al. Development of an efficient and precise adenine base editor (ABE) with expanded target range in allotetraploid cotton (Gossypium hirsutum). BMC Biol. 2022, 20, 45.
- Bharat, S.S.; Li, S.Y.; Li, J.Y.; Yan, L.; Xia, L.Q. Base editing in plants: Current status and challenges. Crop J. 2020, 8, 384–395.
- Butt, H.; Rao, G.S.; Sedeek, K.; Aman, R.; Kamel, R.; Mahfouz, M. Engineering herbicide resistance via prime editing in rice. Plant Biotechnol. J. 2020, 18, 2370–2372.
- Molla, K.A.; Sretenovic, S.; Bansal, K.C.; Qi, Y. Precise plant genome editing using base editors and prime editors. Nat. Plants 2021, 7, 1166–1187.
- Nelson, J.W.; Randolph, P.B.; Shen, S.P.; Everette, K.A.; Chen, P.J.; Anzalone, A.V.; An, M.; Newby, G.A.; Chen, J.C.; Hsu, A.; et al. Engineered pegRNAs improve prime editing efficiency. Nat. Biotechnol. 2022, 40, 402–410.
- Li, Z.; Zhang, D.; Xiong, X.; Yan, B.; Xie, W.; Sheen, J.; Li, J.-F. A potent Cas9-derived gene activator for plant and mammalian cells. Nat. Plants 2017, 3, 930–936.
- Zhang, Y.; Yin, C.; Zhang, T.; Li, F.; Yang, W.; Kaminski, R.; Fagan, P.R.; Putatunda, R.; Young, W.-B.; Khalili, K.; et al. CRISPR/gRNA-directed synergistic activation mediator (SAM) induces specific, persistent and robust reactivation of the HIV-1 latent reservoirs. Sci. Rep. 2015, 5, 16277.
- Pan, C.; Wu, X.; Markel, K.; Malzahn, A.A.; Kundagrami, N.; Sretenovic, S.; Zhang, Y.; Cheng, Y.; Shih, P.M.; Qi, Y. CRISPR–Act3.0 for highly efficient multiplexed gene activation in plants. Nat. Plants 2021, 7, 942–953.
- Ding, X.; Yu, L.; Chen, L.; Li, Y.; Zhang, J.; Sheng, H.; Ren, Z.; Li, Y.; Yu, X.; Jin, S.; et al. Recent Progress and Future Prospect of CRISPR/Cas-Derived Transcription Activation (CRISPRa) System in Plants. Cells 2022, 11, 3045.
- Park, J.J.; Dempewolf, E.; Zhang, W.Z.; Wang, Z.Y. RNA-guided transcriptional activation via CRISPR/dCas9 mimics overexpression phenotypes in Arabidopsis. PLoS ONE 2017, 12, e0179410.
- Xu, Y.; Lin, Q.; Li, X.; Wang, F.; Chen, Z.; Wang, J.; Li, W.; Fan, F.; Tao, Y.; Jiang, Y.; et al. Fine-tuning the amylose content of rice by precise base editing of the Wx gene. Plant Biotechnol. J. 2021, 19, 11–13.
- Tang, Y.; Abdelrahman, M.; Li, J.; Wang, F.; Ji, Z.; Qi, H.; Wang, C.; Zhao, K. CRISPR/Cas9 induces exon skipping that facilitates development of fragrant rice. Plant Biotechnol. J. 2021, 19, 642–644.
- Dong, L.; Qi, X.; Zhu, J.; Liu, C.; Zhang, X.; Cheng, B.; Mao, L.; Xie, C. Supersweet and waxy: Meeting the diverse demands for specialty maize by genome editing. Plant Biotechnol. J. 2019, 17, 1853–1855.
- Wang, Y.; Liu, X.; Zheng, X.; Wang, W.; Yin, X.; Liu, H.; Ma, C.; Niu, X.; Zhu, J.K.; Wang, F. Creation of aromatic maize by CRISPR/Cas. J. Integr. Plant Biol. 2021, 63, 1664–1670.
- Liu, L.; Gallagher, J.; Arevalo, E.D.; Chen, R.; Skopelitis, T.; Wu, Q.; Bartlett, M.; Jackson, D. Enhancing grain-yield-related traits by CRISPR-Cas9 promoter editing of maize CLE genes. Nat. Plants 2021, 7, 287–294.
- Chen, Y.; Fu, M.; Li, H.; Wang, L.; Liu, R.; Liu, Z.; Zhang, X.; Jin, S. High-oleic acid content, nontransgenic allotetraploid cotton (Gossypium hirsutum L.) generated by knockout of GhFAD2 genes with CRISPR/Cas9 system. Plant Biotechnol. J. 2021, 19, 424–426.
- Hassan, M.M.; Yuan, G.; Chen, J.-G.; Tuskan, G.A.; Yang, X. Prime Editing Technology and Its Prospects for Future Applications in Plant Biology Research. BioDesign Res. 2020, 2020, 9350905.
- Zhang, Y.W.; Bai, Y.; Wu, G.H.; Zou, S.H.; Chen, Y.F.; Gao, C.X.; Tang, D.Z. Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 2017, 91, 714–724.
- Li, Y.M.; Zhu, J.J.; Wu, H.; Liu, C.L.; Huang, C.L.; Lan, J.H.; Zhao, Y.M.; Xie, C.X. Precise base editing of non-allelic acetolactate synthase genes confers sulfonylurea herbicide resistance in maize. Crop J. 2020, 8, 449–456.
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015, 33, 41–52.
- Ma, X.L.; Zhu, Q.L.; Chen, Y.L.; Liu, Y.G. CRISPR/Cas9 Platforms for Genome Editing in Plants: Developments and Applications. Mol. Plant 2016, 9, 961–974.
- Wang, S.Y.; Yang, Y.H.; Guo, M.; Zhong, C.Y.; Yan, C.J.; Sun, S.Y. Targeted mutagenesis of amino acid transporter genes for rice quality improvement using the CRISPR/Cas9 system. Crop J. 2020, 8, 457–464.
- Christopoulou, M.; Wo, S.R.C.; Kozik, A.; McHale, L.K.; Truco, M.J.; Wroblewski, T.; Michelmore, R.W. Genome-Wide Architecture of Disease Resistance Genes in Lettuce. G3-Genes Genomes Genet. 2015, 5, 2655–2669.
- Miklis, M.; Consonni, C.; Bhat, R.A.; Lipka, V.; Schulze-Lefert, P.; Panstruga, R. Barley MLO modulates actin-dependent and actin-independent antifungal defense pathways at the cell periphery. Plant Physiol. 2007, 144, 1132–1143.
- Li, S.; Lin, D.; Zhang, Y.; Deng, M.; Chen, Y.; Lv, B.; Li, B.; Lei, Y.; Wang, Y.; Zhao, L.; et al. Genome-edited powdery mildew resistance in wheat without growth penalties. Nature 2022, 602, 455–460.
- Panstruga, R.; Schulze-Lefert, P. Live and let live: Insights into powdery mildew disease and resistance. Mol. Plant Pathol. 2002, 3, 495–502.
- van Esse, H.P.; Reuber, T.L.; van der Does, D. Genetic modification to improve disease resistance in crops. New Phytol. 2020, 225, 70–86.
- Arora, L.; Narula, A. Gene Editing and Crop Improvement Using CRISPR-Cas9 System. Front. Plant Sci. 2017, 8, 1932.
- Silva, C.J.; van den Abeele, C.; Ortega-Salazar, I.; Papin, V.; Adaskaveg, J.A.; Wang, D.; Casteel, C.L.; Seymour, G.B.; Blanco-Ulate, B. Host susceptibility factors render ripe tomato fruit vulnerable to fungal disease despite active immune responses. J. Exp. Bot. 2021, 72, 2696–2709.
- Jeon, J.E.; Kim, J.G.; Fischer, C.R.; Mehta, N.; Dufour-Schroif, C.; Wemmer, K.; Mudgett, M.B.; Sattely, E. A Pathogen-Responsive Gene Cluster for Highly Modified Fatty Acids in Tomato. Cell 2020, 180, 176–187.e119.
- Wang, Y.P.; Cheng, X.; Shan, Q.W.; Zhang, Y.; Liu, J.X.; Gao, C.X.; Qiu, J.L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 2014, 32, 947–951.
- Pramanik, D.; Shelake, R.M.; Park, J.; Kim, M.J.; Hwang, I.; Park, Y.; Kim, J.Y. CRISPR/Cas9-Mediated Generation of Pathogen-Resistant Tomato against Tomato Yellow Leaf Curl Virus and Powdery Mildew. Int. J. Mol. Sci. 2021, 22, 1878.
- Nekrasov, V.; Wang, C.M.; Win, J.; Lanz, C.; Weigel, D.; Kamoun, S. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 2017, 7, 482.
- Malnoy, M.; Viola, R.; Jung, M.H.; Koo, O.J.; Kim, S.; Kim, J.S.; Velasco, R.; Kanchiswamy, C.N. DNA-Free Genetically Edited Grapevine and Apple Protoplast Using CRISPR/Cas9 Ribonucleoproteins. Front. Plant Sci. 2016, 7, 1904.
- Wan, D.Y.; Guo, Y.; Cheng, Y.; Hu, Y.; Xiao, S.; Wang, Y.; Wen, Y.Q. CRISPR/Cas9-mediated mutagenesis of VvMLO3 results in enhanced resistance to powdery mildew in grapevine (Vitis vinifera). Hortic. Res. 2020, 7, 116.
- Liu, Y.; Zeng, J.M.; Yuan, C.; Guo, Y.S.; Yu, H.Q.; Li, Y.P.; Huang, C.J. Cas9-PF, an early flowering and visual selection marker system, enhances the frequency of editing event occurrence and expedites the isolation of genome-edited and transgene-free plants. Plant Biotechnol. J. 2019, 17, 1191–1193.
- Acevedo-Garcia, J.; Kusch, S.; Panstruga, R. Magical mystery tour: MLO proteins in plant immunity and beyond. New Phytol. 2014, 204, 273–281.
- Piffanelli, P.; Ramsay, L.; Waugh, R.; Benabdelmouna, A.; D’Hont, A.; Hollricher, K.; Jorgensen, J.H.; Schulze-Lefert, P.; Panstruga, R. A barley cultivation-associated polymorphism conveys resistance to powdery mildew. Nature 2004, 430, 887–891.
- Consonni, C.; Humphry, M.E.; Hartmann, H.A.; Livaja, M.; Durner, J.; Westphal, L.; Vogel, J.; Lipka, V.; Kemmerling, B.; Schulze-Lefert, P.; et al. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 2006, 38, 716–720.
- Bai, X.D.; Correa, V.R.; Toruno, T.Y.; Ammar, E.D.; Kamoun, S.; Hogenhout, S.A. AY-WB Phytoplasma Secretes a Protein That Targets Plant Cell Nuclei. Mol. Plant-Microbe Interact. 2009, 22, 18–30.
- Pessina, S.; Lenzi, L.; Perazzolli, M.; Campa, M.; Dalla Costa, L.; Urso, S.; Vale, G.; Salamini, F.; Velasco, R.; Malnoy, M. Knockdown of MLO genes reduces susceptibility to powdery mildew in grapevine. Hortic. Res. 2016, 3, 16016.
- Wang, W.; Pan, Q.L.; He, F.; Akhunova, A.; Chao, S.M.; Trick, H.; Akhunov, E. Transgenerational CRISPR-Cas9 Activity Facilitates Multiplex Gene Editing in Allopolyploid Wheat. CRISPR J. 2018, 1, 65–74.
- Fister, A.S.; Landherr, L.; Maximova, S.N.; Guiltinan, M.J. Transient Expression of CRISPR/Cas9 Machinery Targeting TcNPR3 Enhances Defense Response in Theobroma cacao. Front. Plant Sci. 2018, 9, 268.
- Wang, W.; Xue, Z.; Miao, J.; Cai, M.; Zhang, C.; Li, T.; Zhang, B.; Tyler, B.M.; Liu, X. PcMuORP1, an Oxathiapiprolin-Resistance Gene, Functions as a Novel Selection Marker for Phytophthora Transformation and CRISPR/Cas9 Mediated Genome Editing. Front. Microbiol. 2019, 10, 2402.
- Ma, J.; Chen, J.; Wang, M.; Ren, Y.L.; Wang, S.; Lei, C.L.; Cheng, Z.J. Sodmergen, Disruption of OsSEC3A increases the content of salicylic acid and induces plant defense responses in rice. J. Exp. Bot. 2018, 69, 1051–1064.
- Wang, F.J.; Wang, C.L.; Liu, P.Q.; Lei, C.L.; Hao, W.; Gao, Y.; Liu, Y.G.; Zhao, K.J. Enhanced Rice Blast Resistance by CRISPR/Cas9-Targeted Mutagenesis of the ERF Transcription Factor Gene OsERF922. PLoS ONE 2016, 11, e0154027.
- Zhou, Y.; Xu, S.; Jiang, N.; Zhao, X.; Bai, Z.; Liu, J.; Yao, W.; Tang, Q.; Xiao, G.; Lv, C.; et al. Engineering of rice varieties with enhanced resistances to both blast and bacterial blight diseases via CRISPR/Cas9. Plant Biotechnol. J. 2022, 20, 876–885.
- Roossinck, M.J.; Martin, D.P.; Roumagnac, P. Plant Virus Metagenomics: Advances in Virus Discovery. Phytopathology 2015, 105, 716–727.
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013, 11, 777–788.
- Fondong, V.N. Geminivirus protein structure and function. Mol. Plant Pathol. 2013, 14, 635–649.
- Zaidi, S.S.E.A.; Tashkandi, M.; Mansoor, S.; Mahfouz, M.M. Engineering Plant Immunity: Using CRISPR/Cas9 to Generate Virus Resistance. Front. Plant Sci. 2016, 7, 1673.
- Kis, A.; Hamar, E.; Tholt, G.; Ban, R.; Havelda, Z. Creating highly efficient resistance against wheat dwarf virus in barley by employing CRISPR/Cas9 system. Plant Biotechnol. J. 2019, 17, 1004–1006.
- Gilbertson, R.L.; Batuman, O.; Webster, C.G.; Adkins, S. Role of the Insect Supervectors Bemisia tabaci and Frankliniella occidentalis in the Emergence and Global Spread of Plant Viruses. Annu. Rev. Virol. 2015, 2, 67–93.
- Baltes, N.J.; Hummel, A.W.; Konecna, E.; Cegan, R.; Bruns, A.N.; Bisaro, D.M.; Voytas, D.F. Conferring resistance to geminiviruses with the CRISPR-Cas prokaryotic immune system. Nat. Plants 2015, 1, 15145.
- Ji, X.; Zhang, H.; Zhang, Y.; Wang, Y.; Gao, C. Establishing a CRISPR-Cas-like immune system conferring DNA virus resistance in plants. Nat. Plants 2015, 1, 15144.
- Ali, Z.; Abulfaraj, A.; Idris, A.; Ali, S.; Tashkandi, M.; Mahfouz, M.M. CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 2015, 16, 238.
- Ali, Z.; Ali, S.; Tashkandi, M.; Zaidi, S.S.E.A.; Mahfouz, M.M. CRISPR/Cas9-Mediated Immunity to Geminiviruses: Differential Interference and Evasion. Sci. Rep. 2016, 6, 26912.
- Yu, Y.; Pan, Z.; Wang, X.; Bian, X.; Wang, W.; Liang, Q.; Kou, M.; Ji, H.; Li, Y.; Ma, D.; et al. Targeting of SPCSV-RNase3 via CRISPR-Cas13 confers resistance against sweet potato virus disease. Mol. Plant Pathol. 2022, 23, 104–117.
- Zhang, T.; Zheng, Q.F.; Yi, X.; An, H.; Zhao, Y.L.; Ma, S.Q.; Zhou, G.H. Establishing RNA virus resistance in plants by harnessing CRISPR immune system. Plant Biotechnol. J. 2018, 16, 1415–1423.
- Larson, M.H.; Gilbert, L.A.; Wang, X.W.; Lim, W.A.; Weissman, J.S.; Qi, L.S. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 2013, 8, 2180–2196.
- Aman, R.; Ali, Z.; Butt, H.; Mahas, A.; Aljedaani, F.; Khan, M.Z.; Ding, S.W.; Mahfouz, M. RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 2018, 19, 1.
- Zhan, X.; Zhang, F.; Zhong, Z.; Chen, R.; Wang, Y.; Chang, L.; Bock, R.; Nie, B.; Zhang, J. Generation of virus-resistant potato plants by RNA genome targeting. Plant Biotechnol. J. 2019, 17, 1814–1822.
- Noureen, A.; Zuhaib Khan, M.; Amin, I.; Zainab, T.; Ahmad, N.; Haider, S.; Mansoor, S. Broad-spectrum resistance against multiple PVY-strains by CRSIPR/Cas13 system in Solanum tuberosum crop. GM Crops Food 2022, 13, 97–111.
- Chandrasekaran, J.; Brumin, M.; Wolf, D.; Leibman, D.; Klap, C.; Pearlsman, M.; Sherman, A.; Arazi, T.; Gal-On, A. Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 2016, 17, 1140–1153.
- Pyott, D.E.; Sheehan, E.; Molnar, A. Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol. Plant Pathol. 2016, 17, 1276–1288.
- Macovei, A.; Sevilla, N.R.; Cantos, C.; Jonson, G.B.; Slamet-Loedin, I.; Cermak, T.; Voytas, D.F.; Choi, I.R.; Chadha-Mohanty, P. Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant Biotechnol. J. 2018, 16, 1918–1927.
- Sanfacon, H. Plant Translation Factors and Virus Resistance. Viruses 2015, 7, 3392–3419.
- Noureen, A.; Khan, M.Z.; Amin, I.; Zainab, T.; Mansoor, S. CRISPR/Cas9-Mediated Targeting of Susceptibility Factor eIF4E-Enhanced Resistance Against Potato Virus Y. Front. Genet. 2022, 13, 922019.
- Lucioli, A.; Tavazza, R.; Baima, S.; Fatyol, K.; Burgyan, J.; Tavazza, M. CRISPR-Cas9 Targeting of the eIF4E1 Gene Extends the Potato Virus Y Resistance Spectrum of the Solanum tuberosum L. cv. Desirée. Front. Microbiol. 2022, 13, 873930.
- Schloss, P.D.; Girard, R.A.; Martin, T.; Edwards, J.; Thrash, J.C. Status of the Archaeal and Bacterial Census: An Update. mBio 2016, 7, e00201-16.
- Zhou, J.H.; Peng, Z.; Long, J.Y.; Sosso, D.; Liu, B.; Eom, J.S.; Huang, S.; Liu, S.Z.; Cruz, C.V.; Frommer, W.B.; et al. Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 2015, 82, 632–643.
- Oliva, R.; Ji, C.H.; Atienza-Grande, G.; Huguet-Tapia, J.C.; Perez-Quintero, A.; Li, T.; Eom, J.S.; Li, C.H.; Nguyen, H.; Liu, B.; et al. Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 2019, 37, 1344–1350.
- Li, C.; Li, W.; Zhou, Z.; Chen, H.; Xie, C.; Lin, Y. A new rice breeding method: CRISPR/Cas9 system editing of the Xa13 promoter to cultivate transgene-free bacterial blight-resistant rice. Plant Biotechnol. J. 2020, 18, 313–315.
- Langner, T.; Kamoun, S.; Belhaj, K. CRISPR Crops: Plant Genome Editing Toward Disease Resistance. Annu. Rev. Phytopathol. 2018, 56, 479–512.
- Thomazella, D.P.T.; Seong, K.; Mackelprang, R.; Dahlbeck, D.; Geng, Y.; Gill, U.S.; Qi, T.; Pham, J.; Giuseppe, P.; Lee, C.Y.; et al. Loss of function of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. USA 2021, 118, e2026152118.
- Paula de Toledo Thomazella, D.; Brail, Q.; Dahlbeck, D.; Staskawicz, B.J. CRISPR-Cas9 mediated mutagenesis of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. bioRxiv 2016, 064824.
- Gimenez-Ibanez, S.; Boter, M.; Ortigosa, A.; Garcia-Casado, G.; Chini, A.; Lewsey, M.G.; Ecker, J.R.; Ntoukakis, V.; Solano, R. JAZ2 controls stomata dynamics during bacterial invasion. New Phytol. 2017, 213, 1378–1392.
- Ortigosa, A.; Gimenez-Ibanez, S.; Leonhardt, N.; Solano, R. Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of SlJAZ2. Plant Biotechnol. J. 2019, 17, 665–673.
- Sun, L.F.; Nasrullah; Ke, F.Z.; Nie, Z.P.; Wang, P.; Xu, J.G. Citrus Genetic Engineering for Disease Resistance: Past, Present and Future. Int. J. Mol. Sci. 2019, 20, 5256.
- Jia, H.G.; Wang, N. Xcc-facilitated agroinfiltration of citrus leaves: A tool for rapid functional analysis of transgenes in citrus leaves. Plant Cell Rep. 2014, 33, 1993–2001.
- Jia, H.; Orbovic, V.; Jones, J.B.; Wang, N. Modification of the PthA4 effector binding elements in Type I CsLOB1 promoter using Cas9/sgRNA to produce transgenic Duncan grapefruit alleviating XccΔpthA4:dCsLOB1.3 infection. Plant Biotechnol. J. 2016, 14, 1291–1301.
- Peng, A.H.; Chen, S.C.; Lei, T.G.; Xu, L.Z.; He, Y.R.; Wu, L.; Yao, L.X.; Zou, X.P. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 2017, 15, 1509–1519.
- Wang, L.; Chen, S.; Peng, A.; Xie, Z.; He, Y.; Zou, X. CRISPR/Cas9-mediated editing of CsWRKY22 reduces susceptibility to Xanthomonas citri subsp. citri in Wanjincheng orange (Citrus sinensis (L.) Osbeck). Plant Biotechnol. Rep. 2019, 13, 501–510.
- Pompili, V.; Dalla Costa, L.; Piazza, S.; Pindo, M.; Malnoy, M. Reduced fire blight susceptibility in apple cultivars using a high-efficiency CRISPR/Cas9-FLP/FRT-based gene editing system. Plant Biotechnol. J. 2020, 18, 845–858.
This entry is offline, you can click here to edit this entry!
