The mammalian target of rapamycin (mTOR) is the major controller of a number of important cellular activities, including protein synthesis, cell expansion, multiplication, autophagy, lysosomal function, and cellular metabolism. The mTOR signaling system regulates gene transcription and protein manufacturing to control proliferation of cell, differentiation of immune cell, and tumor metabolism. Due to its vital role in case of microbial infections, inflammations and cancer development and progression, mTOR has been considered as a key therapeutic target for the development of targeted medication.
- mammalian target of rapamycin (mTOR)
- infection
1. mTOR Inhibitors
| Sr. No. | Name of mTOR Inhibitor | Therapeutic Importance | Reference |
|---|---|---|---|
| 1 | Rapamycin (Sirolimus) |
|
[10] [11] [12] [13] |
| 2 | Temsirolimus | Treat advanced Renal cancer | [7] |
| 3 | Everolimus |
|
[14] [15] [16] |
| 4 | Ridaforolimus/Deforolimus |
|
[17] [18] |
| 5 | Zotarolimus | Antitumor activity | [19] |
| 6 | Torin 1 | Suppress colon cancer cells | [20] |
| 7 | Torin 2 | Antitumor activity | [21] |
| 8 | MLN0128 | Advanced solid tumors | [22] |
| 9 | AZD2014 (Vistusertib) | Metastatic clear cell renal cancer | [22] |
| 10 | Voxtalisib (SAR24540; XL765) | non-Hodgkin lymphoma or chronic lymphocytic lymphoma that has relapsed or is refractory | [22] |
| 11 | Gedatolisib (PKI-587 PF05212384) | Recurrent endometrial cancer | [22] |
| 12 | Rapalink-1 |
|
[7] |
| 13 | Halitulin analog ICSN3250 | It has the ability to compete with and interchange phospholipids acid in the mTOR FRB domain | [23] |
| 14 | LY3023414 |
|
[24] |
| 15 | O SU-53 |
|
[25] |
| 16 | OSI-027 | Anticancer | [26] |
| 17 | C C-223 | Anticancer | [27] |
| 18 | PKI-587 | Gastroenteropancreatic Neuroendocrine tumor disease | [28] |
| 19 | INK-128 | Inhibit angiogenesis and tumor growth in
|
[29] |
| 20 | GSK2126458 | Robust activity in cancer models | [30] |
| 21 | XL765 | Glioblastoma development is inhibited by triggering ER stress-dependent apoptosis. | [31] |
| 22 | NVP-BEZ235 | Cancer cell proliferation is inhibited by this compound | [32] |
| 23 | P529 | Stops cancer cells from multiplying | [33] |
| 24 | JR-AB2-011 | Anti-glioblastoma multiforme properties | [34] |
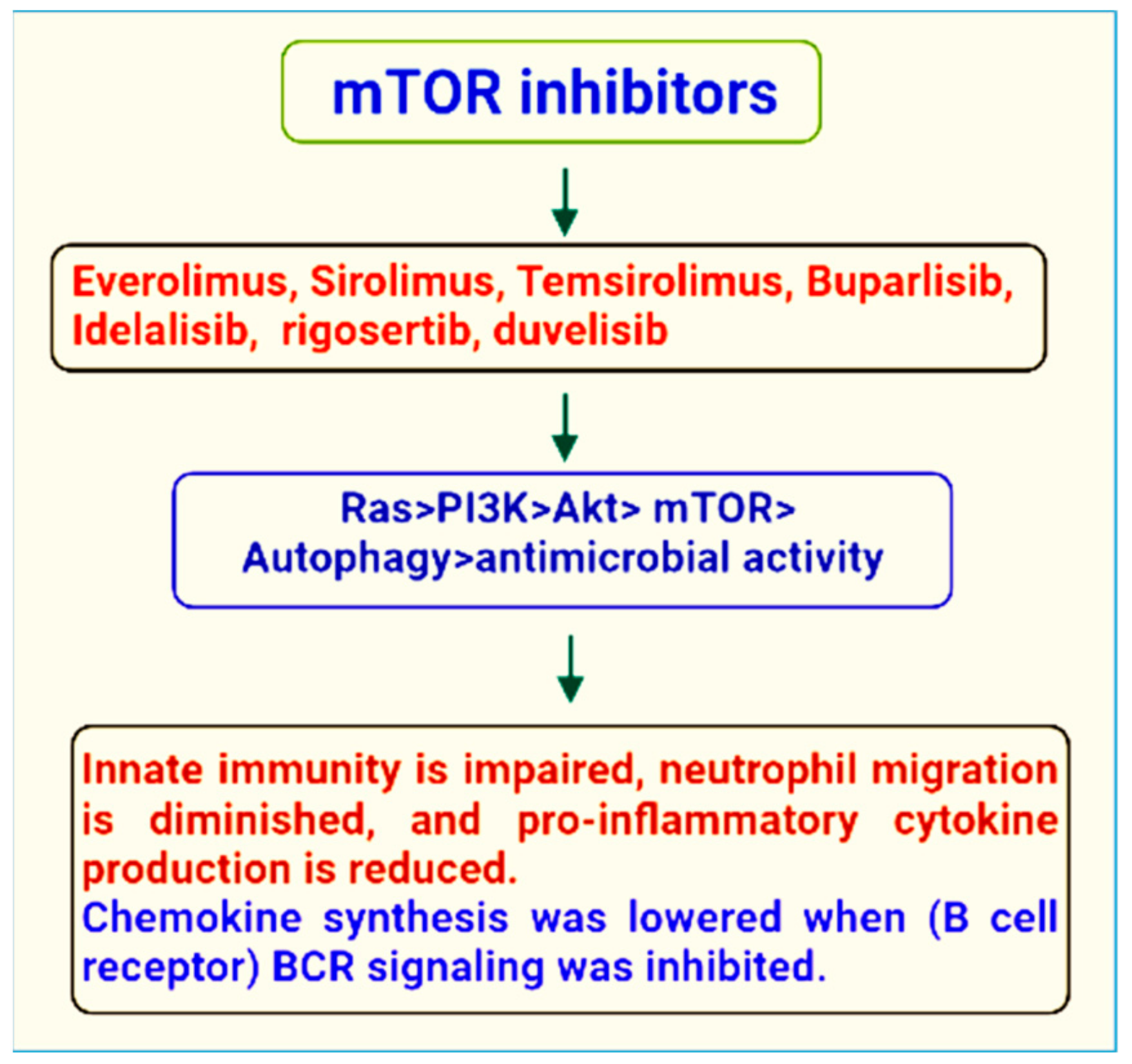
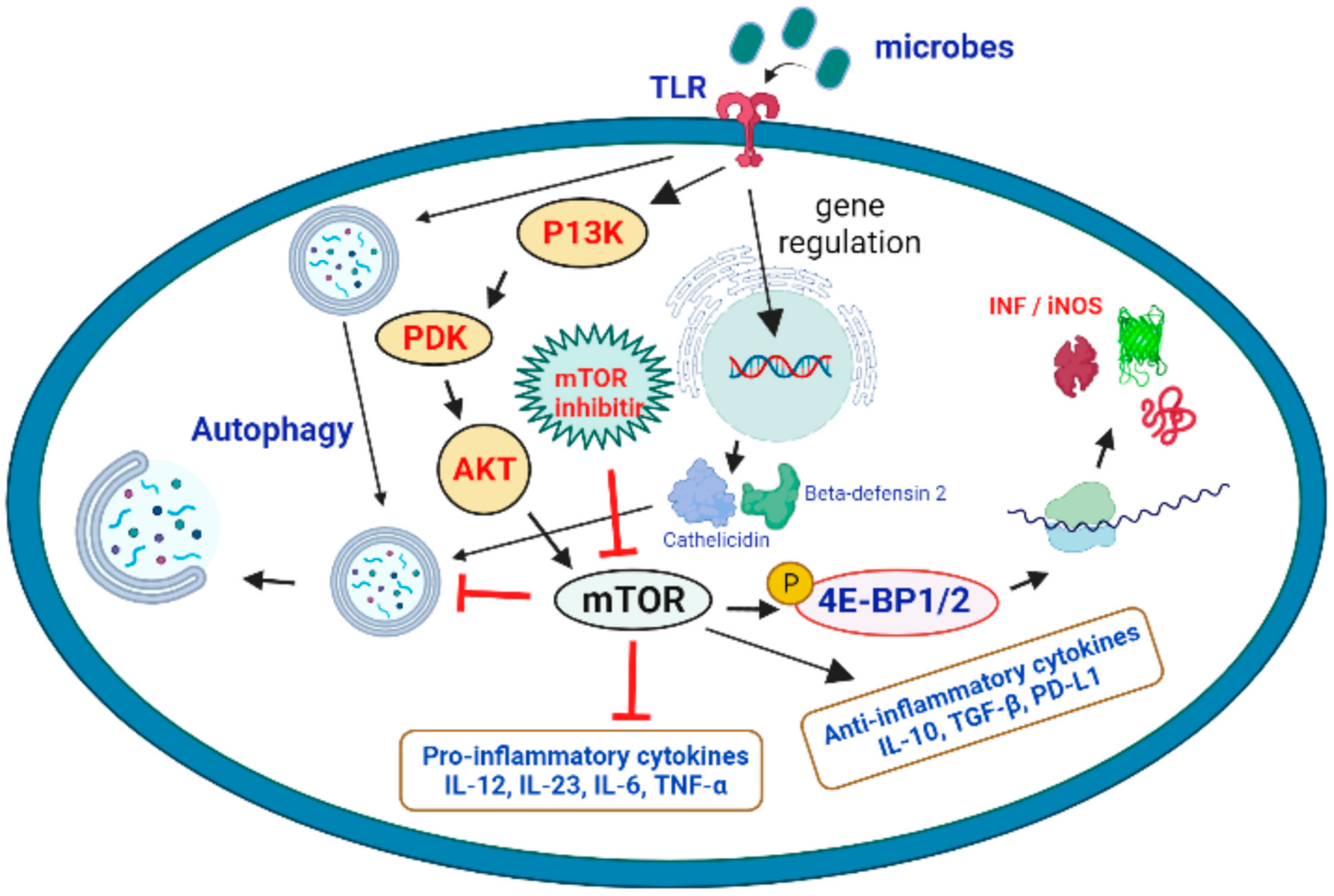
2. Therapeutic Role of mTOR Signaling in the Treatment of Microbial Infections
This entry is adapted from the peer-reviewed paper 10.3390/ijms232012470
References
- Pieri, M.; Miraglia, N.; Polichetti, G.; Tarantino, G.; Acampora, A.; Capone, D. Analytical and Pharmacological Aspects of Therapeutic Drug Monitoring of mTOR Inhibitors. Curr. Drug Metab. 2011, 12, 253–267.
- Bullock, K.E.; Petros, W.P.; Younis, I.; Uronis, H.E.; Morse, M.A.; Blobe, G.C.; Zafar, S.Y.; Gockerman, J.P.; Lager, J.J.; Truax, R.; et al. A phase I study of bevacizumab (B) in combination with everolimus (E) and erlotinib (E) in advanced cancer (BEE). Cancer Chemother. Pharmacol. 2011, 67, 465–474.
- Capone, D.; Palmiero, G.; Gentile, A.; Basile, V.; Federico, S.; Sabbatini, M.; Potenza, M.; Perfetti, A.; Pieri, M.; Tarantino, G. A Pharmacokinetic Interaction Between Clarithromycin and Sirolimus in Kidney Transplant Recipient. Curr. Drug Metab. 2007, 8, 379–381.
- Palavra, F.; Robalo, C.; Reis, F. Recent Advances and Challenges of mTOR Inhibitors Use in the Treatment of Patients with Tuberous Sclerosis Complex. Oxidative Med. Cell. Longev. 2017, 2017, 1–11.
- Zhou, H.-Y.; Huang, S.-L. Current development of the second generation of mTOR inhibitors as anticancer agents. Chin. J. Cancer 2013, 32, 8–18.
- Popova, N.V.; Jücker, M. The Role of mTOR Signaling as a Therapeutic Target in Cancer. Int. J. Mol. Sci. 2021, 22, 1743.
- Rodrik-Outmezguine, V.S.; Okaniwa, M.; Yao, Z.; Novotny, C.J.; McWhirter, C.; Banaji, A.; Won, H.; Wong, W.; Berger, M.; De Stanchina, E.; et al. Overcoming mTOR resistance mutations with a new-generation mTOR inhibitor. Nature 2016, 534, 272–276.
- Powers, R.W.; Kaeberlein, M.; Caldwell, S.D.; Kennedy, B.K.; Fields, S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006, 20, 174–184.
- Granata, S.; Gassa, A.D.; Carraro, A.; Brunelli, M.; Stallone, G.; Lupo, A.; Zaza, G. Sirolimus and Everolimus Pathway: Reviewing Candidate Genes Influencing Their Intracellular Effects. Int. J. Mol. Sci. 2016, 17, 735.
- Pal, S.K.; Quinn, D.I. Differentiating mTOR inhibitors in renal cell carcinoma. Cancer Treat. Rev. 2013, 39, 709–719.
- Malagelada, C.; Jin, Z.H.; Jackson-Lewis, V.; Przedborski, S.; Greene, L.A. Rapamycin Protects against Neuron Death in In Vitro andIn Vivo Models of Parkinson’s Disease. J. Neurosci. 2010, 30, 1166–1175.
- Ma, K.L.; Ruan, X.Z.; Powis, S.H.; Moorhead, J.F.; Varghese, Z. Anti-atherosclerotic effects of sirolimus on human vascular smooth muscle cells. Am. J. Physiol. Circ. Physiol. 2007, 292, H2721–H2728.
- Goudar, R.K.; Shi, Q.; Hjelmeland, M.D.; Keir, S.T.; McLendon, R.E.; Wikstrand, C.J.; Reese, E.D.; Conrad, C.A.; Traxler, P.; Lane, H.A.; et al. Combination therapy of inhibitors of epidermal growth factor receptor/vascular endothelial growth factor receptor 2 (AEE788) and the mammalian target of rapamycin (RAD001) offers improved glioblastoma tumor growth inhibition. Mol. Cancer Ther. 2005, 4, 101–112.
- Kirchner, G.I.; Meier-Wiedenbach, I.; Manns, M.P. Clinical Pharmacokinetics of Everolimus. Clin. Pharmacokinet. 2004, 43, 83–95.
- Kovarik, M.; Everolimus, A. Proliferation signal inhibitor targeting primary causes of allograft dysfunction. Drugs Today 2004, 40, 101–109.
- Yao, J.C.; Shah, M.H.; Ito, T.; Bohas, C.L.; Wolin, E.M.; Van Cutsem, E.; Hobday, T.J.; Okusaka, T.; Capdevila, J.; de Vries, E.G.; et al. Everolimus for Advanced Pancreatic Neuroendocrine Tumors. N. Engl. J. Med. 2011, 364, 514–523.
- Sessa, C.; Tosi, D.; Viganò, L.; Albanell, J.; Hess, D.; Maur, M.; Cresta, S.; Locatelli, A.; Angst, R.; Rojo, F.; et al. Phase Ib study of weekly mammalian target of rapamycin inhibitor ridaforolimus (AP23573; MK-8669) with weekly paclitaxel. Ann. Oncol. 2009, 21, 1315–1322.
- Chawla, S.P.; Staddon, A.P.; Baker, L.H.; Schuetze, S.M.; Tolcher, A.W.; D’Amato, G.Z.; Blay, J.-Y.; Mita, M.M.; Sankhala, K.K.; Berk, L.; et al. Phase II Study of the Mammalian Target of Rapamycin Inhibitor Ridaforolimus in Patients with Advanced Bone and Soft Tissue Sarcomas. J. Clin. Oncol. 2020, 30, 78–84, Coriggendum in 2017, 35, 2722.
- Serruys, P.W.; Silber, S.; Garg, S.; van Geuns, R.J.; Richardt, G.; Buszman, P.E.; Kelbæk, H.; van Boven, A.J.; Hofma, S.H.; Linke, A.; et al. Comparison of Zotarolimus-Eluting and Everolimus-Eluting Coronary Stents. N. Engl. J. Med. 2010, 363, 136–146.
- Francipane, M.G.; Lagasse, E. mTOR pathway in colorectal cancer: An update. Oncotarget 2014, 5, 49–66.
- Liu, Q.; Wang, J.; Kang, S.A.; Thoreen, C.C.; Hur, W.; Ahmed, T.; Sabatini, D.M.; Gray, N.S. Discovery of 9-(6-Aminopyridin-3-yl)-1-(3-(trifluoromethyl)phenyl)benzo1,6naphthyridin-2(1H)-one (Torin2) as a Potent, Selective, and Orally Available Mammalian Target of Rapamycin (mTOR) Inhibitor for Treatment of Cancer. J. Med. Chem. 2011, 54, 1473–1480.
- Hua, H.; Kong, Q.; Zhang, H.; Wang, J.; Luo, T.; Jiang, Y. Targeting mTOR for cancer therapy. J. Hematol. Oncol. 2019, 12, 71.
- Nguyen, T.-L.; Nokin, M.-J.; Egorov, M.; Tomé, M.; Bodineau, C.; Di Primo, C.; Minder, L.; Wdzieczak-Bakala, J.; Garcia-Alvarez, M.C.; Bignon, J.; et al. mTOR Inhibition via Displacement of Phosphatidic Acid Induces Enhanced Cytotoxicity Specifically in Cancer Cells. Cancer Res. 2018, 78, 5384–5397.
- Bendell, J.C.; Varghese, A.M.; Hyman, D.M.; Bauer, T.M.; Pant, S.; Callies, S.; Lin, J.; Martinez, R.; Wickremsinhe, E.R.; Fink, A.; et al. A First-in-Human Phase 1 Study of LY3023414, an Oral PI3K/mTOR Dual Inhibitor, in Patients with Advanced Cancer. Clin. Cancer Res. 2018, 24, 3253–3262.
- Plews, R.L.; Yusof, A.M.; Wang, C.; Saji, M.; Zhang, X.; Chen, C.-S.; Ringel, M.D.; Phay, J.E. A Novel Dual AMPK Activator/mTOR Inhibitor Inhibits Thyroid Cancer Cell Growth. J. Clin. Endocrinol. Metab. 2015, 100, E748–E756.
- Lee, D.-F.; Kuo, H.-P.; Chen, C.-T.; Hsu, J.-M.; Chou, C.-K.; Wei, Y.; Sun, H.-L.; Li, L.-Y.; Ping, B.; Huang, W.-C.; et al. IKKβ Suppression of TSC1 Links Inflammation and Tumor Angiogenesis via the mTOR Pathway. Cell 2007, 130, 440–455.
- von Manteuffel, S.R.; Gingras, A.C.; Ming, X.F.; Sonenberg, N.; Thomas, G. 4E-BP1 phosphorylation is mediated by the FRAP-p70s6k pathway and is independent of mitogen-activated protein kinase. Proc. Natl. Acad. Sci. USA 1996, 93, 4076–4080.
- Yip, C.K.; Murata, K.; Walz, T.; Sabatini, D.M.; Kang, S.A. Structure of the Human mTOR Complex I and Its Implications for Rapamycin Inhibition. Mol. Cell 2010, 38, 768–774.
- Saucedo, L.J.; Gao, X.; Chiarelli, D.A.; Li, L.; Pan, D.; Edgar, B.A. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat. Cell Biol. 2003, 5, 566–571.
- Ma, L.; Chen, Z.; Erdjument-Bromage, H.; Tempst, P.; Pandolfi, P.P. Phosphorylation and Functional Inactivation of TSC2 by Erk: Implications for Tuberous Sclerosisand Cancer Pathogenesis. Cell 2005, 121, 179–193.
- Inoki, K.; Ouyang, H.; Zhu, T.; Lindvall, C.; Wang, Y.; Zhang, X.; Yang, Q.; Bennett, C.; Harada, Y.; Stankunas, K.; et al. TSC2 Integrates Wnt and Energy Signals via a Coordinated Phosphorylation by AMPK and GSK3 to Regulate Cell Growth. Cell 2006, 126, 955–968.
- Hardie, D.G. AMP-activated/SNF1 protein kinases: Conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 2007, 8, 774–785.
- Shahbazian, D.; Roux, P.; Mieulet, V.; Cohen, M.S.; Raught, B.; Taunton, J.; Hershey, J.W.B.; Blenis, J.; Pende, M.; Sonenberg, N. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J. 2006, 25, 2781–2791.
- Lenz, G.; Avruch, J. Glutamatergic Regulation of the p70S6 Kinase in Primary Mouse Neurons. J. Biol. Chem. 2005, 280, 38121–38124.
- Keating, R.; McGargill, M.A. mTOR regulation of lymphoid cells in immunity to pathogens. Front. Immunol. 2016, 7, 180.
- Parate, S.; Kumar, V.; Lee, G.; Rampogu, S.; Hong, J.; Lee, K. Marine-Derived Natural Products as ATP-Competitive mTOR Kinase Inhibitors for Cancer Therapeutics. Pharmaceuticals 2021, 14, 282.
- Yordy, B.; Iwasaki, A. Autophagy in the control and pathogenesis of viral infection. Curr. Opin. Virol. 2011, 1, 196–203.
- Siqueira, M.d.S.; Ribeiro, R.d.M.; Travassos, L.H. Autophagy and its interaction with intracellular bacterial pathogens. Front. Immunol. 2018, 9, 935.
- Bahia, D.; Satoskar, A.R.; Dussurget, O. Cell signaling in host–pathogen interactions: The host point of view. Front. Immunol. 2018, 9, 221.
- Jostins, L.; Ripke, S.; Weersma, R.K.; Duerr, R.H.; McGovern, D.P.; Hui, K.Y.; Lee, J.C.; Schumm, L.P.; Sharma, Y.; Anderson, C.A.; et al. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012, 491, 119–124.
- Axtell, R.C.; De Jong, B.A.; Boniface, K.; Van Der Voort, L.F.; Bhat, R.; De Sarno, P.; Naves, R.; Han, M.; Zhong, F.; Castellanos, J.G.; et al. T helper type 1 and 17 cells determine efficacy of interferon-β in multiple sclerosis and experimental encephalomyelitis. Nat. Med. 2010, 16, 406–412.
- Pollizzi, K.N.; Powell, J.D. Regulation of T cells by mTOR: The known knowns and the known unknowns. Trends Immunol. 2014, 36, 13–20.
- Limon, J.J.; Fruman, D.A. Akt and mTOR in B Cell Activation and Differentiation. Front. Immunol. 2012, 3, 228.
- Babiński, S.; Giermaziak, H. Influenza epidemic in 1971 in diabetics treated with 1-butyl-biguanidine hydrochloride (Silubin retard) and 1-phenylethyl-biguanidine hydrochloride (Phenformin). Polski Tyg. Lek. 1973, 28, 1815–1817.
- Chuang, Y.-C.; Ruan, S.-Y. Compelling Results of Adjuvant Therapy with Sirolimus for Severe H1N1 Pneumonia. Crit. Care Med. 2014, 42, e687–e688.
- Liu, L.; Das, S.; Losert, W.; Parent, C.A. mTORC2 regulates neutrophil chemotaxis in a cAMP-and RhoA-dependent fashion. Dev. Cell 2010, 19, 845–857.
- Yang, H.; Wang, X.; Zhang, Y.; Liu, H.; Liao, J.; Shao, K.; Chu, Y.; Liu, G. Modulation of TSC-mTOR signaling on immune cells in immunity and autoimmunity. J. Cell. Physiol. 2014, 229, 17–26.
- Squarize, C.; Castilho, R.M.; Bugge, T.H.; Gutkind, J.S. Accelerated Wound Healing by mTOR Activation in Genetically Defined Mouse Models. PLoS ONE 2010, 5, e10643.
- Katholnig, K.; Linke, M.; Pham, H.; Hengstschläger, M.; Weichhart, T. Immune responses of macrophages and dendritic cells regulated by mTOR signalling. Biochem. Soc. Trans. 2013, 41, 927–933.
- Kufer, T.A.; Banks, D.J.; Philpott, D.J. Innate Immune Sensing of Microbes by Nod Proteins. Ann. N. Y. Acad. Sci. 2006, 1072, 19–27.
- Abdel-Nour, M.; Tsalikis, J.; Kleinman, D.; Girardin, S.E. The emerging role of mTOR signalling in antibacterial immunity. Immunol. Cell Biol. 2014, 92, 346–353.
- Rampersad, S.; Tennant, P. Replication and Expression Strategies of Viruses. Viruses 2018, 55–82.
- Le Sage, V.; Cinti, A.; Amorim, R.; Mouland, A.J. Adapting the Stress Response: Viral Subversion of the mTOR Signaling Pathway. Viruses 2016, 8, 152.
- Terrazzano, G.; Rubino, V.; Palatucci, A.T.; Giovazzino, A.; Carriero, F.; Ruggiero, G. An open question: Is it rational to inhibit the mTor-dependent pathway as COVID-19 therapy? Front. Pharmacol. 2020, 11, 856.
- Castle, B.T.; Dock, C.; Hemmat, M.; Kline, S.; Tignanelli, C.; Rajasingham, R.; Masopust, D.; Provenzano, P.; Langlois, R.; Schacker, T.; et al. Biophysical modeling of the SARS-CoV-2 viral cycle reveals ideal antiviral targets. Biorxiv 2020.
- Beatman, E.; Oyer, R.; Shives, K.D.; Hedman, K.; Brault, A.C.; Tyler, K.L.; Beckham, J.D. West Nile virus growth is independent of autophagy activation. Virology 2012, 433, 262–272.
- Cheng, F.; da Silva, S.R.; Huang, I.-C.; Jung, J.U.; Gao, S.-J. Suppression of Zika Virus Infection and Replication in Endothelial Cells and Astrocytes by PKA Inhibitor PKI 14-22. J. Virol. 2018, 92, e02019-17.
- Nakashima, K.; Takeuchi, K.; Chihara, K.; Hotta, H.; Sada, K. Inhibition of hepatitis C virus replication through adenosine monophosphate-activated protein kinase-dependent and-independent pathways. Microbiol. Immunol. 2011, 55, 774–782.
- Del Campo, J.A.; Garcia-Valdecasas, M.; Gil-Gomez, A.; Rojas, A.; Gallego, P.; Ampuero, J.; Gallego-Durán, R.; Pastor, H.; Grande, L.; Padillo, F.J.; et al. Simvastatin and metformin inhibit cell growth in hepatitis C virus infected cells via mTOR increasing PTEN and autophagy. PLoS ONE 2018, 13, e0191805.
- Proud, C.G. Phosphorylation and Signal Transduction Pathways in Translational Control. Cold Spring Harb. Perspect. Biol. 2019, 11, a033050.
- Wang, C.-H.; Chung, F.-T.; Lin, S.-M.; Huang, S.-Y.; Chou, C.-L.; Lee, K.-Y.; Lin, T.-Y.; Kuo, H.-P. Adjuvant Treatment With a Mammalian Target of Rapamycin Inhibitor, Sirolimus, and Steroids Improves Outcomes in Patients With Severe H1N1 Pneumonia and Acute Respiratory Failure*. Crit. Care Med. 2014, 42, 313–321.
- Denys, A.; Bocian, J. Effect of Silubin-retard (1-butyl-biguanide hydrochloride) on the course of influenza-virus infection in mice. Polski Tyg. Lek. 1970, 25, 332–334.
- Rightsel, W.A.; Dice, J.R.; McAlpine, R.J.; Timm, E.A.; McLean, I.W.; Dixon, G.J.; Schabel, F.M. Antiviral Effect of Guanidine. Science 1961, 134, 558–559.
- Ueda, T.; Toyoshima, S.; Tsuji, T.; Seto, Y.; Nomoto, J. Antiviral effect of guanidine and its derivatives part 1. the inhibitory effect of guanidine on the multiplication of poliomyelitis virus in tissue culture. Keio J. Med. 1961, 10, 257–265.
- Loddo, B. Inhibition of the multiplication in vitro of poliomyelitis viruses by guanidine. VIII. Elective inhibitory activity of guanidine on the PCE of enteroviruses. Boll. Della Soc. Ital. Di Biol. Sper. 1962, 38, 8–10.
- Loddo, B.; Ferrari, W.; Brotzu, G.; Spanedda, A. Inhibitory action of guanidine on the multiplication of enteroviruses and especially of polioviruses. Boll. Dell’Istituto Sieroter. Milan. 1962, 41, 111–120.
- Kindrachuk, J.; Ork, B.; Hart, B.J.; Mazur, S.; Holbrook, M.R.; Frieman, M.B.; Traynor, D.; Johnson, R.F.; Dyall, J.; Kuhn, J.H.; et al. Antiviral Potential of ERK/MAPK and PI3K/AKT/mTOR Signaling Modulation for Middle East Respiratory Syndrome Coronavirus Infection as Identified by Temporal Kinome Analysis. Antimicrob. Agents Chemother. 2015, 59, 1088–1099.
- Garcia, G., Jr.; Sharma, A.; Ramaiah, A.; Sen, C.; Kohn, D.; Gomperts, B.; Svendsen, C.N.; Damoiseaux, R.D.; Arumugaswami, V. Antiviral drug screen of kinase inhibitors identifies cellular signaling pathways critical for SARS-CoV-2 replication. BioRxiv 2020.
- Betz, C.; Hall, M.N. Where is mTOR and what is it doing there? J. Cell Biol. 2013, 203, 563–574.
- Appelberg, S.; Gupta, S.; Svensson Akusjärvi, S.; Ambikan, A.T.; Mikaeloff, F.; Saccon, E.; Végvári, Á.; Benfeitas, R.; Sperk, M.; Ståhlberg, M. Dysregulation in Akt/mTOR/HIF-1 signaling identified by proteo-transcriptomics of SARS-CoV-2 infected cells. Emerg. Microbes Infect. 2020, 9, 1748–1760.
- Luo, P.; Qiu, L.; Liu, Y.; Liu, X.-L.; Zheng, J.-L.; Xue, H.-Y.; Liu, W.-H.; Liu, D.; Li, J. Metformin Treatment Was Associated with Decreased Mortality in COVID-19 Patients with Diabetes in a Retrospective Analysis. Am. J. Trop. Med. Hyg. 2020, 103, 69–72.
