The flexibility of worldwide climate changes every year, global warming, waterlogging, drought, salinity, pest, and the pathogen, impedes crop productivity. Brassica napus is one of the most important oil crops in the world, and rapeseed oil is considered one of the conducive edible vegetable oil to health. Recently, miRNAs have been found and identified to control the expression of targets under disruptive environmental conditions. The mechanism is through the formation of the silencing complex that mediates post-transcriptional gene silencing, which pairs the target mRNA and target cleavage and/or translation inhibition. However, the functional role of miRNAs and targets in B. napus is still not clarified. This review focuses on the current knowledge of miRNAs concerning development regulation, biotic and abiotic stress responses in B. napus. Moreover, more strategies for miRNAs manipulation in plants are discussed, along with future perspectives, and the enormous amount of transcriptome data available provides cues for miRNAs functions in B. napus. Finally, the construction miRNA regulatory network and development of climate change tolerance B. napus is significance.
- miRNA
- Brassica napus
- development regulation
- biotic stress
- abiotic stress
- transcriptome
1. Introduction
Like abiotic stress, biotic stress including viruses, bacteria, fungi, insect pests, and nematode parasites has also affected on the growth and development of plants [68]. miRNAs have been identified that involved in the regulation of biotic stress and immune response in plants. There are many common diseases in plants, and different plants are infected with different diseases. In Arabidopsis, A total of 293 known miRNAs and 6 potential novel sRNAs were identified from 15 small RNA libraries in post-inoculation leaves with Phytophthora capsici (P. capsici) using high-throughput sequencing [69]. miR38-3P, a novel miRNA, was highly induced in expression after infection of the pathogen Sclerotinia sclerotiorum, which might target AT3G03820 in the involvement of Arabidopsis-Sclerotinia interaction [70]. To enhance the ability of Arabidopsis against pathogen infection, Bacillus velezensis FZB42-treated library and control library were constructed, 11 known miRNAs and 4 novel miRNAs were differentially expressed after FZB42 inoculation [71]. These results showed that miRNAs and its target have closely associated with defense response. In wheat, small RNA high-throughput sequencing was used to screen and identify miRNAs involved in powdery mildew stress response. The results showed that 24 miRNAs might be involved in powdery mildew stress response, among which 8 miRNAs responded to powdery mildew infection in susceptible wheat cultivar Jingdong8 (JD8). miR2001, miR2006 and miR2011 were down-regulated after powdery mildew infection, and miR393, miR444, miR827, miR2005, and miR2013 were up-regulated. Three miRNAs responded to powdery mildew infection in JD8-Pm30, a near-isogenic resistant line of JD8, including miR171 down-regulated and miR2008 and miR2012 up-regulated after powdery mildew infection. There were 10 miRNAs that responded to powdery mildew infection in both JD8 and JD8-Pm30, among which miR156, miR159, miR164, and miR396 were down-regulated after powdery mildew infection [72]. In tomato, a total 79 plant miRNAs and 40 potential candidate miRNAs were differentially expressed after Cucumber mosaic virus (CMV)-infection [73]. The fungus Magnaporthe oryzae (M. oryzae) is the most important disease in rice, the expression level of rice miR319 was induced by M. oryzae strain Guy11. miR319 and its target gene TEOSINTE BRANCHED/CYCLOIDEA/PROLIFERATING CELL FACTOR1 (OsTCP21) may participate in the process of blast M. oryzae [74]. In addition, previous study showed suppressing the expression of miR482 and increasing the level of NBS (nucleotide binding site)-LRR (leucine-rich repeat) transcript could increase the resistance of cotton to Verticillium dahliae [75]. miR482 and its target genes NBS-LRR is involved in regulating potato resistance against Verticillium dahliae infection in potato [76]. Moreover, miR472a could also target NBS-LRRs is involved in effective defence against the necrotrophic fungus Cytospora chrysosperma in poplar[77].
2. miRNAs and development regulation in B. napus
| Functions | MicroRNAs | References |
|---|---|---|
| Branch angle regulation | Multiple miRNAs | [90,91] |
| Flower development | miR172 | [92] |
| Male sterility | miR159 | [93] |
| Silique development | miR160, miR2111, miR399, miR827, and miR408 | [95] |
| Thickness of pod canopy | miR159, miR6029, and miR827 | [96] |
| Seed development | Multiple miRNAs | [94,97,100] |
| Fatty acid and content | Multiple miRNAs | [98,99,101,102] |
Review
Multiple functions of miRNAs in Brassica napus L.
Jian Li 1 ,Yangyang Li 1 ,Rongyuan Wang 1 ,Jiangyan Fu 1 , Xinxing Zhou 1 ,Yujie Fang 2 ,Youping Wang 2 and Yaju Liu 1,*
|
Citation: Lastname, F.; Lastname, F.; Lastname, F. Title. Life 2022, 12, x. https://doi.org/10.3390/xxxxx Academic Editor: Firstname Lastname Received: date Accepted: date Published: date Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Copyright: © 2022 by the authors. Submitted for possible open access publication under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/). |
1 Xuzhou Institute of Agricultural Sciences in Jiangsu Xuhuai District, 221121, Xuzhou, China
2 Key Laboratory of Plant Functional Genomics of the Ministry of Education/Jiangsu Key Laboratory of Crop Genomics and Molecular Breeding, Yangzhou University, 225009, Yangzhou, China
* Correspondence: yajuliu@jaas.ac.cn (Y. L.); Tel.: 86+516-82189969
Simple Summary: Changing climatic conditions are posing serious challenges to crop production. In B. napus, the vast amount of transcriptomic data on development, biotic and abiotic stress offer one possibility to exploit the regulatory network of miRNA-mRNA. This review summarizes the advances of miRNA-related regulation under disruptive environmental conditions and learns more about the characteristics of miRNAs for improvement of favorable traits in B. napus.
Abstract: The flexibility of worldwide climate changes every year, global warming, waterlogging, drought, salinity, pest, and the pathogen, impedes crop productivity. Brassica napus is one of the most important oil crops in the world, and rapeseed oil is considered one of the conducive edible vegetable oil to health. Recently, miRNAs have been found and identified to control the expression of targets under disruptive environmental conditions. The mechanism is through the formation of the silencing complex that mediates post-transcriptional gene silencing, which pairs the target mRNA and target cleavage and/or translation inhibition. However, the functional role of miRNAs and targets in B. napus is still not clarified. This review focuses on the current knowledge of miRNAs concerning development regulation, biotic and abiotic stress responses in B. napus. Moreover, more strategies for miRNAs manipulation in plants are discussed, along with future perspectives, and the enormous amount of transcriptome data available provides cues for miRNAs functions in B. napus. Finally, the construction miRNA regulatory network and development of climate change tolerance B. napus is significance.
Keywords: miRNA; Brassica napus; development regulation; biotic stress; abiotic stress; transcriptome
1. Introduction
Amphidiploid Brassica napus L. (B. napus L.) is the third oilseed crop after soybean and palm. It is widely planted and distributed in the world, and plays a vital role in vegetable oil, biofuel, and livestock feeding [1]. Nowadays, the rapeseed planting area of China ranks the first in the world, but the total rapeseed production is still unable to meet the market demand with the increasing population and disruptive environmental conditions [2, 3]. Biotic stresses (such as microbial infections) and abiotic stresses (such as drought, heat, flooding, salinity, etc.) are frequent and disruptive environmental conditions, various stresses will reduce the growth of biomass and root system, leaf number, specific leaf area, photosynthesis, and chlorophyll content in B. napus, while stresses at flowering or silique stage may lead to earlier flowering time and lower seed weight, oil content and fatty acid content, which greatly restrict the growth and development of rapeseed, and ultimately affect the yield and quality of rapeseed, and endanger food safety worldwide [4]. Therefore, improving the yield of rapeseed and deciphering the mechanism of rapeseed against various stresses are the most important strategies to meet the increasing edible oil demand [5, 6].
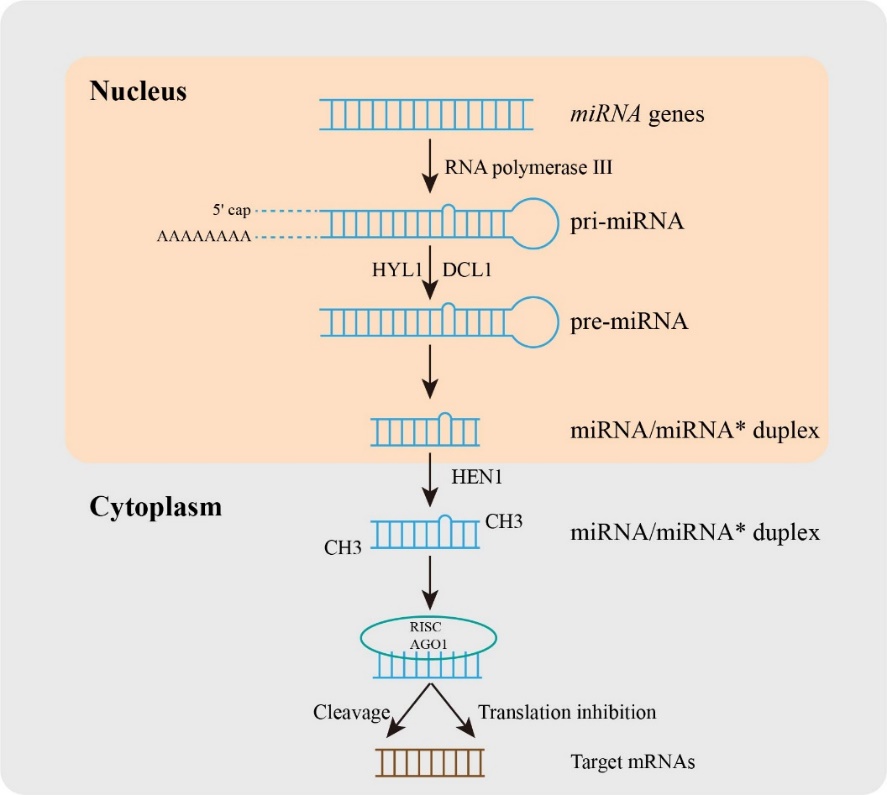
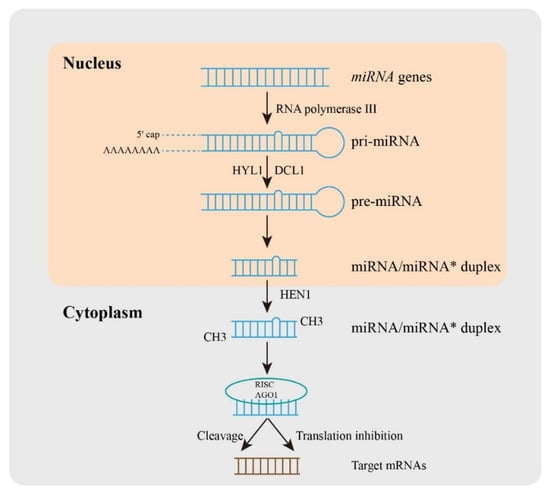
Small RNAs (sRNAs) are almost 20-30 nucleotides of non-coding RNA, including small interfering RNAs (siRNAs) [7], Piwi-interacting RNAs (piRNAs) [8], and microRNAs (miRNAs) [9]. The single-stranded miRNAs as key regulators are known to control the expression of target mRNAs and participate in the regulation of normal plant growth [10], development [11], as well as biotic and abiotic stress responses [12]. The first non-coding RNA was found in Caenorhabditis elegans in 1993 [13], and then Thomas Tuschl, David Bartel, and Victor Ambros laboratories used miRNA to name these small RNAs in published articles in 2001 [14]. Over the decades, emerging research on miRNAs identification and characterization has given a new method for plant species improvement. After that, Sanger developed the miRBase (http://www.mirbase.org/) database and established naming rules and usage specifications for miRNAs [15]. Then, the comprehensive and detailed database of small RNAs was built in plants including miRFANs [16], TarDB [17], or sRNAanno [18]. In plants (Figure 1), the miRNA biosynthesis process is the transcription of miRNA genes (MIRs) under the action of RNA polymerase III to produce the primary miRNA (pri-miRNA), and the stem-loop structure is formed through 5' caping and 3' polyadenylation of long pri-miRNA in the nucleus [19]. The precursor miRNA (pre-miRNA) with stem-loop structure was formed by the cleavage complex DICER like1 (DCL1) [20], HYPONASTIC LEAVES1 (HYL1) [21], and SERRATE (SE) [22], and then pre-miRNA was cut into double-stranded miRNA/miRNA* under the action of the cleavage complex [23]. The transporters carried it from the nucleus to the cytoplasm, and its 3' end was methylated under the action of methyltransferase HUA-ENHANCER1 (HEN1), and finally formed a double-stranded stable mature miRNA/miRNA * [19]. The mature miRNA is then loaded into the RNA-induced silencing complex (RISC), and regulates gene expression. The regulation of miRNA is mainly based on the principle that the seed region of miRNA mature sequence near-perfectly matches the sequence of the target gene mRNA [24]. RISC recognizes the target region and combines it with the target region mediated by Argonaute (AGO) protein to change the expression of the target gene and thus affect the physiological process of plants. The regulation of miRNA on target genes in plants is mainly through two modes including target cleavage and/or translation inhibition [25]. Moreover, different miRNAs may interact with the same targets, at the same time, one miRNA may regulate different targets.
|
|
Figure 1. The processes of miRNA biogenesis in plants. In brief, a miRNA gene was transcribed into primary miRNA (pri-miRNA) with the help of RNA polymerase III, and then pri-miRNA was produced by the cleavage complex DICER like1 (DCL1), HYPONASTIC LEAVES1 (HYL1), and other proteins. The pre-miRNA is cleaved into double-stranded miRNA/miRNA*. The transporter carries it from the nucleus to the cytoplasm, and its 3' end is methylated under the action of methyltransferase HUA-ENHANCER1 (HEN1), which eventually forms double-stranded stable mature miRNA/miRNA*. Mature miRNAs are loaded into RNA-induced silencing complex (RISC) and regulate gene expression.
miRNAs have been shown to be involved in plant growth and development through various signaling pathways, indicating that these miRNAs can function as developmental signaling molecules in plants [26]. Studies have reported that inhibition of DCL1 and the HASTY expression of important proteins in the process of miRNA biosynthesis in plants will reduce the abundance of miRNA expression, seriously affect morphology, resulting in changes in leaf shape and flower shape, pollination obstruction, fertility reduction, etc. [27-29]. miR160 negatively regulates ARF10 to maintain the homeostasis of ARF10-mediated interactions between auxin and ABA pathways during seed germination and postembryonic development [30]. In Arabidopsis, miR395c negatively regulates seed germination under high salinity or dehydration stress, miR395e were only single nucleotide differences to miR395c. However, miR395c and miR395e act as positive or negative regulators of seed germination under stress conditions [31]. miRNA controls leaf development by regulating the expression of HD-ZIP transcription factors [32]. As important members of the HD-ZIP transcription factor family, including PHB, PHV, and REV [33], miR165 can regulates leaf development by controlling the expression of these three target genes [34-36]. In rice, miR167 negatively regulates the expression of several auxin response factor genes (ARF8 and ARF6), and further affects the expression of IAA-binding enzyme gene OSGH3-2, participating in the regulation of exogenous auxin and determining the content of intracellular beneficial auxin [37]. In Arabidopsis, miR396 inhibits cell proliferation during leaf development by inhibiting the expression of its target gene GRF and cell cycle-related genes [38]. The TCP gene of Arabidopsis is the target gene of miR159. TCP genes in several plant species have miRNA binding sites, indicating that miRNA-mediated regulation of leaf morphogenesis has a conserved role in plants with different leaf shapes [39]. miR156 and miR172 play a key role in the process of vegetative leaf development at the late germination stage, and play an important role in the transition of plants to growth stage [40]. Studies in maize have found that miR172 negatively regulates the number of leaves in maize during vegetative growth by controlling the expression of glossy15 (GL15) [41]. Ectopic expression of apple Md-miR156h in Arabidopsis inhibits the expression of SPL family members SPL17 and SPL19, thereby delaying plant flowering, indicating that miR156 mediates a conserved post-transcriptional regulatory pathway in apple and Arabidopsis [42]. miR164c can negatively regulate the expression of transcription factors CUC1 and CUC2, increasing the petals of Arabidopsis. At the same time, it was found that similar members of the same miRNA family targeting the same group of genes would play different functions due to different expression patterns during development [43], e.g. miR172 promotes flowering and destroys floral organ characteristics by down-regulating the expression of target gene APETALA2 (AP2) [44]. In rice, studies have shown that overexpression of miR172 can cause spikelets deletion and floral organ deformity, and miR172b plays a role in florets development by regulating the expression of target gene APETALA2-like [45]. miR172 repression of EAT3 (TOE3) is essential for floral organogenesis in Arabidopsis. In addition, SPL3 targeted by miR156 can directly activate the expression of TOE3, indicating a novel signal interaction between miR156 and miR172 in the process of flower organ formation [46]. Leaf senescence controlled by the age of plant development and aggravated by environmental stresses such as drought, high temperature and salinity [47]. Overexpressing SlymiR208 in tomato significantly induced early leaf senescence phenotype in SlIPT4 gene silencing transgenic plants, indicating that SlymiR208 positively regulates leaf senescence in tomato mainly by regulating SlIPT2 and SlIPT4 related to cytokinin synthesis [48]. ORESARA1 (ORE1) is a key senescence regulator in Arabidopsis thaliana, and miR164 is involved in the regulation of leaf senescence by inhibiting ORE1 gene expression at the post-transcriptional level [49, 50]. These findings indicate that miRNAs play an important role in plant development, participating in the regulation of seed germination, stem, leaf, flower and other different organ development.
miRNA-mediated post-transcriptional regulation has been shown to be involved in plant responses to a variety of abiotic stresses [51]. To identify miRNAs and their target genes under drought stress in peach and almond trees, qPCR was used to analyze the expression levels of miR156, miR159, miR160, miR167, and miR171 under moderate and severe water shortage conditions [52]. miR166 can improve drought resistance of rice by causing morphological changes such as leaf curl and xylem diameter reduction [53]. Lateral root growth of transgenic rice seedlings overexpressing TIR1 and AFB2 resistant to miR393-cleaved forms was no longer inhibited by ABA or osmotic stress. This indicates that miR393-mediated attenuation of auxin signal can regulate the adaptation of plant roots to drought stress [54]. Overexpression of OsmiR393 and OsmiR393b in rice could improve the sensitivity of transgenic rice to salt stress, overexpression of OsmiR393 in Arabidopsis also has the same phenotype [55]. Wheat TaMIR1119 plays an important role in regulating plant drought tolerance by regulating plant osmotic accumulation, photosynthesis and improving ROS homeostasis in cells [56]. The highly conserved miR156/SPL module plays an important role in balancing plant growth and stress response. In Tamarix chinensis, the miR156/SPL module plays a regulatory function in mediating the response to salt stress [57]. miRNA is also involved in the regulation of plant response to extreme environmental temperature. In sunflower, miR396 responses to heat stress by regulating the expression of target gene HaWRKY6 [58]. In Arabidopsis, low temperature can induce the up-regulation of miR393 and miR319c [59]. Overexpression of miR397a can affect the expression level of COR gene downstream of cold tolerance gene CBF, improving the tolerance of transgenic plants to low temperature [60]. As a key factor of cold stress induction, miR319 is induced by cold stress in a variety of plants. The response of 12 miRNAs in sugarcane to cold stress identified the differentially expressed miR319 under normal conditions and low temperature stress [61]. Subsequently, 18 cold-responsive miRNAs were identified using microarray in rice, and most of them were found to be down-regulated with cold induction [62]. Overexpressing OsmiR319b had increased proline content, increased survival rate and significantly increased resistance to low temperature [63]. It has also been found that the expression levels of SlymiR166 and SlymiR319 in tomato was increased under cold stress conditions [64]. Other abiotic stresses, including oxidative stress and nutrient stress such as nitrogen and phosphorus deficiency, also seriously restrict plant growth. A total of 144 miRNAs related to hydrogen peroxide (H2O2) stress were identified by next-generation sequencing technology combined with qPCR and 5' RACE analysis in Brachypodium distachyon, and their target genes were analyzed, revealing the response and defense mechanism to oxidative stress at the post-transcriptional regulatory level [65]. In addition, the phosphoric acid transporter NtPT2 gene was up-regulated in TamiR408 overexpressing plants, overexpressing TamiR408 showed stronger stress tolerance, higher biomass and photosynthate under low phosphorus conditions [66]. The expression level of Arabidopsis miR167a is significantly increased under low nitrogen stress, which can affect the lateral root growth under low nitrogen stress by targeting ARF6 and ARF8 [67].
Like abiotic stress, biotic stress including viruses, bacteria, fungi, insect pests, and nematode parasites has also affected on the growth and development of plants [68]. miRNAs have been identified that involved in the regulation of biotic stress and immune response in plants. There are many common diseases in plants, and different plants are infected with different diseases. In Arabidopsis, A total of 293 known miRNAs and 6 potential novel sRNAs were identified from 15 small RNA libraries in post-inoculation leaves with Phytophthora capsici (P. capsici) using high-throughput sequencing [69]. miR38-3P, a novel miRNA, was highly induced in expression after infection of the pathogen Sclerotinia sclerotiorum, which might target AT3G03820 in the involvement of Arabidopsis-Sclerotinia interaction [70]. To enhance the ability of Arabidopsis against pathogen infection, Bacillus velezensis FZB42-treated library and control library were constructed, 11 known miRNAs and 4 novel miRNAs were differentially expressed after FZB42 inoculation [71]. These results showed that miRNAs and its target have closely associated with defense response. In wheat, small RNA high-throughput sequencing was used to screen and identify miRNAs involved in powdery mildew stress response. The results showed that 24 miRNAs might be involved in powdery mildew stress response, among which 8 miRNAs responded to powdery mildew infection in susceptible wheat cultivar Jingdong8 (JD8). miR2001, miR2006 and miR2011 were down-regulated after powdery mildew infection, and miR393, miR444, miR827, miR2005, and miR2013 were up-regulated. Three miRNAs responded to powdery mildew infection in JD8-Pm30, a near-isogenic resistant line of JD8, including miR171 down-regulated and miR2008 and miR2012 up-regulated after powdery mildew infection. There were 10 miRNAs that responded to powdery mildew infection in both JD8 and JD8-Pm30, among which miR156, miR159, miR164, and miR396 were down-regulated after powdery mildew infection [72]. In tomato, a total 79 plant miRNAs and 40 potential candidate miRNAs were differentially expressed after Cucumber mosaic virus (CMV)-infection [73]. The fungus Magnaporthe oryzae (M. oryzae) is the most important disease in rice, the expression level of rice miR319 was induced by M. oryzae strain Guy11. miR319 and its target gene TEOSINTE BRANCHED/CYCLOIDEA/PROLIFERATING CELL FACTOR1 (OsTCP21) may participate in the process of blast M. oryzae [74]. In addition, previous study showed suppressing the expression of miR482 and increasing the level of NBS (nucleotide binding site)-LRR (leucine-rich repeat) transcript could increase the resistance of cotton to Verticillium dahliae [75]. miR482 and its target genes NBS-LRR is involved in regulating potato resistance against Verticillium dahliae infection in potato [76]. Moreover, miR472a could also target NBS-LRRs is involved in effective defence against the necrotrophic fungus Cytospora chrysosperma in poplar[77].
The miRNAs induced under various stresses can fine-tune the expression of target genes that are functioning in the regulation of stress tolerance in B. napus. Hence, it is necessary to understand miRNA regulation during combat stress conditions. In the present review, we have discussed miRNA regulation in plant development, biotic and abiotic stress responses in B. napus from recent research progress, dissected functional studies to decipher the regulation network behind miRNA-based stress tolerance, and designed stress-resilient rapeseed through the manipulation of miRNAs.
2. miRNAs and development regulation in B. napus
miRNAs in the regulation of plant development have been investigated in diverse plant species, for instance, Arabidopsis [39], rice [78], wheat [79], tomato [80], maize [81], strawberry [82], sugarcane [83], apple [84], sweet potato [85], and ornamental gloxinia [86]. With the rapid development of biotechnology such as high-throughput sequencing, thousands of miRNAs also were identified under rapeseed development [87]. As shown in Table 1, the known miRNAs in Arabidopsis and rice were used to search for potential miRNAs in EST and GSS databases of B. napus [88]. After strict filtering criteria, 21 miRNAs were detected, and 67 potential target genes were further found through blast of the mRNA database [89]. The branch angle determines the planting density of B. napus in the field, and a smaller branch angle can increase the planting density of B. napus, thus improve yield of B. napus. Sequences of two B. napus varieties with different branch angles reveal the relationship between miRNA-related target genes and auxin or BR signaling pathways, which can be finely regulated by changing the expression of these genes in B. napus [90, 91]. The 17 euAP2 genes targeted by miR172 were identified and these genes showed highly expression in floral organs in B. napus, suggesting that miR172-euAP2 may function in flower development [92]. Recently, 12 small RNA libraries of genic male sterility lines in rapeseed were constructed and sequenced to analyze the differential expression of miRNAs in regulating the pollen development, the results showed that miR159 may regulate the fertility in rapeseed [93]. Meanwhile, silique and seed development are also important points to improve the production and quality of rapeseed [94]. Rapeseed genotypes with long and short siliques were used to establish small RNA libraries and 17 differential expressed miRNAs were identified. These miRNAs, such as miR159, miR319, miR160, miR399, miR408, miR827, and miR2111, may be involved in cell proliferation, auxin signal transduction, and inorganic phosphate/copper deficiency to control silique development [95]. Some miRNAs, such as miR159, miR6029, and miR827, were identified to regulate the thickness of pod canopy for yield information [96]. Moreover, more than 500 miRNAs were identified during seed maturation from 10-50 days after flowering in rapeseed using next-generation sequencing, among them, miR156, miR159, miR172, miR167, miR158, and miR166 were found to be involved in the regulation of seed development and maturation [97]. The composition and content of fatty acids affect the quality of rapeseed oil [98, 99]. Computational studies using the high-oil-content and low-oil-content rapeseed cultivars identified some miRNAs that may be involved in regulating the oil content of B. napus [100]. Other studies have also shown that miRNAs play a role in the synthesis of fatty acids, and miRNAs participate in the formation of acetyl-CoA and carbon chain desaturase, regulate the level of long chain fatty acids, β-oxidation, and lipid transport and metabolism, thereby affect the synthesis of fatty acids in B. napus[101, 102]. Therefore, the miRNA regulation of silique development and fatty acid synthesis may have a role in the yield of B. napus, possibly influencing oil content.
Table 1. The functions of miRNAs in B. napus development.
|
Functions |
MicroRNAs |
References |
|
Branch angle regulation |
Multiple miRNAs |
[90, 91] |
|
Flower development |
miR172 |
[92] |
|
Male sterility |
miR159 |
[93] |
|
Silique development |
miR160, miR2111, miR399, miR827, and miR408 |
[95] |
|
Thickness of pod canopy |
miR159, miR6029, and miR827 |
[96] |
|
Seed development |
Multiple miRNAs |
[94, 97, 100] |
|
Fatty acid and content |
Multiple miRNAs |
[98, 99, 101, 102] |
3. miRNAs and abiotic stress in B. napus
Abiotic stress is the most widely studied miRNA-mediated regulation in plant including drought stress [103], salt stress [104], cold stress [105], cadmium stress [106], and nutrient deprivation [107]. Drought and salt stress severely affect the germination of rapeseed [108]. To investigate the regulatory function of miRNAs in the germination of rapeseed under drought and salt stress (Table 2), the rapeseed seeds were exposed to drought and salt treatment, and then the 85 known miRNAs and 882 novel miRNAs were identified by high-throughput sequencing. Among them, miR156, miR169, miR860, miR399, miR171, and miR395 were down-regulated and miR172 was up-regulated under drought- or salt stress [109]. Further, repressing the expression of miR169 improved drought resistance by targeting NF-YA8 in B. napus [110]. Other than drought and salt stress, cold stress has also been studied in rapeseed, a total of 70 known miRNAs and 126 novel miRNAs were identified in leaf tissues under 4°C conditions, and 25 known and 104 novel miRNAs were differentially expressed in rapeseed [111].
Cadmium (Cd) is one of the most toxic heavy metals and with its high mobility in soil, it is easy to be absorbed and accumulated in plants [112, 113]. Excessive accumulation of cadmium in plants will affect plant development and cell function, and sometimes have a fatal impact on plants [114]. A total of 84 miRNAs were identified from four small RNA libraries and identified 802 targets for 37 miRNA families by Cd-treated rapeseed [115]. BnNRAMP1b is regulated by miR167 in rapeseed at the post-transcriptional level. BnNRAMP1b is related to the transportation of intracellular and extracellular environmental substances in B. napus, which can help heavy metal Cd into the rapeseed cell and lead to cell poisoning. The negative regulation of miR167 on BnNRAMP1b can effectively inhibit this process and help rapeseed nullify Cd damage [116]. miR395 and miR158 were also confirmed to play a role in Cd detoxification in B. napus [117, 118]. Overexpression of miR395 increased Cd tolerance in B. napus [117].
Rapeseed growth and seed production need optimal nutrient allocation under sub-optimal conditions [119]. A lot of miRNAs had been identified and characterized from the phloem in rapeseed [120]. A previous study showed that miR399 was induced by phosphate (P) starvation, and miR399 is potentially involved in long-distance communication via the phloem following phosphate deprivation [121]. miR398 and miR395 were up-graded in phloem sap under copper and sulphate starvation respectively [120]. Besides, miRNA microarray results showed that miR395 is also a potential long-distance molecule for transporting via the phloem [122]. miR2111, miR169, and a miR827-like sequence can respond to P and nitrogen (N) status in rapeseed phloem sap [123]. Furthermore, degradome sequencing and RT-qPCR assays revealed that miR827 regulates the process of N-induced leaf senescence, and rapeseed root development under N deficiency depends on the regulation of the miR171-SCL6 and miR160-ARF17 pathways in rapeseed [124]. Taken together, such miRNAs were found to be involved in the regulation of abiotic stress, but little is known about the impact of stress-related miRNAs on their target genes in B. napus. Therefore, miRNAs and targets can become the new targets for designing abiotic stress-resilient rapeseed.
Table 2. The functions of miRNAs in B. napus under biotic and abiotic stresses.
|
Stress |
MicroRNAs |
References |
|
Salt and drought stress |
Multiple miRNAs |
[109, 111] |
|
Drought stress |
miR169 |
[110] |
|
Cadmium stress |
miR158, miR167, miR395 etc. |
[112-118] |
|
Nutrient stress |
miR395, miR398, miR399 etc. |
[120-123] |
|
Vascular disease |
miR168 |
[125] |
|
Sclerotinia rot |
miR159, miR5139, and miR390 etc. |
[126-128] |
|
Clubroot disease |
Multiple miRNAs |
[129] |
4. miRNAs and biotic stress in B. napus
Pathogen invasion, bacterial, and insect are the most usual biotic stress. However, vascular disease and sclerotinia rot are the most destructive diseases in Brassica species that cause significant crop losses every year [130, 131] (Table 2). The fungi spread in the plants by means of hyphal growth or conidia transporting from infected root to shoot [125]. miR393 was the first miRNA regulated plant antibacterial PTI (pattern triggered immunity) through auxin signaling pathway in Arabidopsis [132]. In B. napus, vascular disease is caused by Verticillium longisporum (V. longisporum). A total of 893 B. napus miRNAs including 360 conserved and 533 novel miRNAs were identified from V. longisporum infected/noninfected roots, and miRNA168-AGO1 were associated with compatible plant–V. longisporum interaction [125]. Some miRNAs responsive to Sclerotinia sclerotiorum (S. sclerotiorum) infection have been identified by high-throughput deep sequencing, and their targets were predicted using degradome sequencing to explain the complex mechanism of S. sclerotiorum infection [126, 127]. In addition, the expression of miR159, miR5139, and miR390 altered in response to S. sclerotiorum. A miR1885-triggered disease resistance gene-derived secondary sRNA locus was also identified and verified with degradome sequencing [128]. On top of that, Differential expression of miRNAs was identified in the potential regulation of clubroot disease with Plasmodiophora brassicae [129]. Overall, In the process of pathogen infection, the specific functional role of miRNA in the defense response of fungi needs to be further studied.
5. Discussion and future perspectives
Brassica napus had a large and complex genome by hybridization between Brassica rapa and Brassica oleracea [133]. A variety of natural disasters limited the growth and development of rapeseed to a great extent, affected the yield and quality of rapeseed and endangered the food safety of China. In the face of the different stresses, developing the stress-tolerant rapeseed is one of the most economical and effective methods for biological breeding. With the release of the B. napus genome and high-throughput sequencing technology has been widely applied, the research of B. napus molecular breeding has entered an explosive stage. At present, the psRNATarget database [134] and degradome sequencing [135] are powerful tools to predict and validate the target genes of known miRNAs, and to illuminate the regulatory network of miRNAs and their target genes in the normal development and response to detrimental environment in rapeseed. Although, many miRNAs have been identified based on the next-generation sequencing in rapeseed under different stresses, little is known about the molecular basis of miRNAs in B. napus. Therefore, after the identification of rapeseed miRNA under different stresses, further studies should be focused on the exploration of function, which is essential to develop the stress-tolerant improvement through miRNA manipulation. Alternatively, miRNA may affect the rapeseed development and stress tolerance through various auxin pathways, the crosstalk of miRNA and plant hormone should be verified to expand our knowledge of the role in rapeseed miRNA function, and their associated regulatory networks represent a compelling area of research to pursue in the future. Another respect we should focus on is that one miRNA may have more than one targets, which would cause different effects on rapeseed growth, development, and stress-tolerant. Whether there are common characteristics and functions of the same miRNAs in the regulation of different stresses in rapeseed, and how miRNAs help rapeseed resist stress by regulating target genes, still need to be confirmed by more studies. We should also consider how to select the optimized targets and balance between normal development and improve the tolerance of different stresses aiming to develop the ideal for high resistance and high yield.
There are important directions regarding the interaction between rapeseed and microorganisms for improving yield and stress tolerance. The subterranean microbiota of plants plays a crucial role in plant growth and health, as root-associated microbes can perform important ecological functions. S. sclerotiorum is pathogenic bacteria that widely infects the reproductive growth of rapeseed and causes the loss of rapeseed production. The previous study has identified more miRNAs in response to S. sclerotiorum infection by high-throughput deep sequencing. However, its in-depth analysis to untangle the complex regulatory networks and their cross-talks require further research. Apart from pathogenic bacteria, non-pathogenic bacteria are inescapable functions in plants. For example, the rhizobial endophytes have the ability of nitrogen fixation to promote soybean growth and soybean yield and improve the tolerance of abiotic stress in soybean [136, 137]. In rice, plant growth-promoting bacteria (PGPB) is not only effective in improving rice productivity but also combat bacterial rice pathogens. Applications of PGPB provide an eco-friendly alternative to agroecosystems [138]. Besides, the root bacteria of B. napus were found to can enhance the rapeseed yield [139], and whether those root bacteria may help rapeseed combat the various stresses need to be explored. Meanwhile, we should consider the relationship between miRNA and non-pathogenic bacteria and provide new insight into their cross-talk, filling the gap in the research on the relationship between miRNA and non-pathogenic bacteria in B. napus.
Nowadays, gene transformation technologies have been applied to confer stress-resilient and high-yield capacity in plants including B. napus [140]. In recent researches, the stress-resilient of transgenic plants can be increased by overexpressing miRNA. Meanwhile, inhibition of miRNA activity by target mimicry (MIM) and short tandem target mimic (STTM) technology has been applied in various plants [141, 142]. MIM technology that the non-protein coding gene IPS1, contains a motif with the sequence complementarity to the miRNA, resulting in repression miRNA cleavage [143]. STTM technology can be served as the updated version of MIM for enhancing the inhibition of miRNA activity [144]. In rapeseed, repression of miR169 by target mimicry can impart the tolerance to drought stress [110], but STTM technology has not been used in rapeseed. CRISPR/Cas9 technology is now a very popular method for genome editing. Nowadays, CRISPR/Cas9 can mutant the miRNA genes to uncover their function [145] or edit the miRNAs recognition sites of target genes to change their expression [146]. In rapeseed, a few studies have utilized the CRISPR/Cas9 system to edit genes associated with plant development [147, 148], pod shattering[149, 150], seed production [151], fatty acid composition [152], and responses to various stresses [153]. However, gene editing of miRNAs has not been used in rapeseed, and there are more prospects to develop and use a variety of tools for miRNA manipulation in B. napus. Sometimes, the role of different miRNAs is also a non-negligible relationship, which is worth our consideration and in-depth exploration. Finally, identification of stress-resistance related miRNAs and construction of the plant miRNA network in development or stress resistance improvement through miRNA manipulation in B. napus.
6. Conclusion
With the continuous progress of biotechnology and the reduction of technology costs, the research on miRNA prediction and regulation is more and more extensive in plants. In B. napus, the vast amount of transcriptomic data on development, biotic and abiotic stress offer one possibility to exploit the regulatory network of miRNA-mRNA. The advances in the tools and the sequence information from related plants also provide a reference for more unknown miRNAs in rapeseed, to learn more about the characteristics of miRNAs for improvement of favorable traits in B. napus and provide the basis for the breeding of multiple-resistant rapeseed.
Author Contributions: Finalization of the Review, Y.J.L.; Original Draft Preparation of the Review, J.L., Y.Y.L., R.Y.W.; Editing, J.Y.F., X.X.Z., Y.J.F., Y.P.W. All authors have read and agreed to the published version of the manuscript.
Funding: Not applicable.
Institutional Review Board Statement: Not applicable.
Informed Consent Statement: Not applicable.
Data Availability Statement: Not applicable.
Conflicts of Interest: The authors declare no conflict of interest.
References
1. Tian C, Zhou X, Liu Q, Peng J, Zhang Z, Song H, Ding Z, Zhran MA, Eissa MA, Kheir AMS, Fahmy AE, Abou-Elwafa SF. Increasing yield, quality and profitability of winter oilseed rape (Brassica napus) under combinations of nutrient levels in fertiliser and planting density. Crop Pasture Sci. 2020, 71, 1010-1019.
2. Fu D, Jiang L, Mason AS, Xiao M, Zhu L, Li L, Zhou Q, Shen C, Huang C. Research progress and strategies for multifunctional rapeseed: a case study of China. J Integr Agr. 2016, 15, 1673-1684.
3. Zhu M, Monroe JG, Suhail Y, Villiers F, Mullen J, Pater D, Hauser F, Jeon BW, Bader JS, Kwak JM, Schroeder JI, McKay JK, Assmann SM. Molecular and systems approaches towards drought-tolerant canola crops. New Phytol. 2016, 210, 1169-1189.
4. Dresselhaus T, Hückelhoven R. Biotic and abiotic stress responses in crop plants. Agron J. 2018, 8, 267.
5. Lohani N, Jain D, Singh MB, Bhalla PL. Engineering multiple abiotic stress tolerance in canola, Brassica napus. Front Plant Sci. 2020, 11, 3.
6. So KKY, Duncan RW. Breeding canola (Brassica napus L.) for protein in feed and food. Plants. 2021, 10, 2220.
7. Guo Z, Wang X, Wang Y, Li W, Gal-On A, Ding S. Identification of a new host factor required for antiviral RNAi and amplification of viral siRNAs. Plant Physiol. 2017, 176, 1587-1597.
8. Wu PH, Zamore PD. Defining the functions of PIWI-interacting RNAs. Nat Rev Mol Cell Bio. 2021, 22, 239-240.
9. Zhang L, Xiang Y, Chen S, Shi M, Jiang X, He Z, Gao S. Mechanisms of microRNA biogenesis and stability control in plants. Front Plant Sci. 2022, 13, 844149.
10. Cui C, Wang J, Zhao J, Fang Y, He X, Guo H, Duan C. A brassica miRNA regulates plant growth and immunity through distinct modes of action. Mol Plant. 2020, 13, 231-245.
11. Begum Y. Regulatory role of microRNAs (miRNAs) in the recent development of abiotic stress tolerance of plants. Gene. 2022, 821, 146283.
12. Chand Jha U, Nayyar H, Mantri N, Siddique KHM. Non-Coding RNAs in legumes: their emerging roles in regulating biotic/abiotic stress responses and plant growth and development. Cells. 2021, 10, 1674.
13. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993, 75, 843-854.
14. Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, Matzke M, Ruvkun G, Tuschl T. A uniform system for microRNA annotation. RNA. 2003, 9, 277-279.
15. Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, 140-144.
16. Liu H, Jin T, Liao R, Wan L, Xu B, Zhou S, Guan J. miRFANs: an integrated database for Arabidopsis thaliana microRNA function annotations. BMC Plant Biol. 2012, 12, 68.
17. Liu J, Liu X, Zhang S, Liang S, Luan W, Ma X. TarDB: an online database for plant miRNA targets and miRNA-triggered phased siRNAs. BMC Genomics. 2021, 22, 348.
18. Chen C, Li J, Feng J, Liu B, Feng L, Yu X, Li G, Zhai J, Meyers BC, Xia R. sRNAanno—a database repository of uniformly annotated small RNAs in plants. Hortic Res. 2021, 8, 45.
19. Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005, 307, 932-935.
20. O'Brien J, Hayder H, Zayed Y, Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol. 2018, 9, 402.
21. Han MH, Goud S, Song L, Fedoroff N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc Natl Acad Sci U S A. 2004, 101, 1093-1098.
22. Machida S, Chen H, Adam Yuan Y. Molecular insights into miRNA processing by Arabidopsis thaliana SERRATE. Nucleic Acids Res. 2011, 39, 7828-7836.
23. Dolata J, Taube M, Bajczyk M, Jarmolowski A, Szweykowska-Kulinska Z, Bielewicz D. Regulation of plant microprocessor function in shaping microRNA landscape. Front Plant Sci. 2018, 9, 753.
24. Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol. 2008, 18, 758-762.
25. Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Gene Dev. 2002, 16, 2313-2313.
26. Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MC. microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature. 2004, 428, 84-88.
27. Liu B, Li P, Li X, Liu C, Cao S, Chu C, Cao X. Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiol. 2005, 139, 296-305.
28. Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol. 2002, 12, 1484-1495.
29. Bollman KM, Aukerman MJ, Park MY, Hunter C, Berardini TZ, Poethig RS. HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development. 2003, 130, 1493-1504.
30. Liu PP, Montgomery TA, Fahlgren N, Kasschau KD, Nonogaki H, Carrington JC. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 2007, 52, 133-146.
31. Kim JY, Lee HJ, Jung HJ, Maruyama K, Suzuki N, Kang H. Overexpression of microRNA395c or 395e affects differently the seed germination of Arabidopsis thaliana under stress conditions. Planta. 2010, 232, 1447-1454.
32. Juarez M, Timmermans M. MiRNAs specify dorsoventral polarity during leaf development2004. 551-552 p.
33. Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol. 2003, 13, 1768-1774.
34. Bao N, Lye KW, Barton MK. MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev Cell. 2004, 7, 653-662.
35. Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5' region. Embo j. 2004, 23, 3356-3364.
36. Williams L, Grigg SP, Xie M, Christensen S, Fletcher JC. Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development. 2005, 132, 3657-3668.
37. Yang JH, Han SJ, Yoon EK, Lee WS. Evidence of an auxin signal pathway, microRNA167-ARF8-GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Res. 2006, 34, 1892-1899.
38. Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development. 2010, 137, 103-112.
39. Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. Control of leaf morphogenesis by microRNAs. Nature. 2003, 425, 257-263.
40. Nonogaki H. MicroRNA gene regulation cascades during early stages of plant development. Plant Cell Physiol. 2010, 51, 1840-1846.
41. Lauter N, Kampani A, Carlson S, Goebel M, Moose SP. microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc Natl Acad Sci U S A. 2005, 102, 9412-9417.
42. Sun C, Zhao Q, Liu D, You C, Hao Y. Ectopic expression of the apple Md-miRNA156h gene regulates flower and fruit development in Arabidopsis. Plant Cell. 2013, 112, 343-351.
43. Baker CC, Sieber P, Wellmer F, Meyerowitz EM. The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr Biol. 2005, 15, 303-315.
44. Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell. 2003, 15, 2730-2741.
45. Zhu QH, Upadhyaya NM, Gubler F, Helliwell CA. Over-expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice (Oryza sativa). BMC Plant Biol. 2009, 9, 149.
46. Jung JH, Lee S, Yun J, Lee M, Park CM. The miR172 target TOE3 represses AGAMOUS expression during Arabidopsis floral patterning. Plant Sci. 2014, 215-216, 29-38.
47. Khanna-Chopra R. Leaf senescence and abiotic stresses share reactive oxygen species-mediated chloroplast degradation. Protoplasma. 2012, 249, 469-481.
48. Zhang Y, Yin S, Tu Y, Mei H, Yang Y. A novel microRNA, SlymiR208, promotes leaf senescence via regulating cytokinin biosynthesis in tomato. Physiol Plant. 2020, 169, 143-155.
49. Li Z, Peng J, Wen X, Guo H. Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell. 2013, 25, 3311-3328.
50. Qiu K, Li Z, Yang Z, Chen J, Wu S, Zhu X, Gao S, Gao J, Ren G, Kuai B, Zhou X. EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis. PLoS Genet. 2015, 11, e1005399.
51. Zhang B, Pan X, Cobb GP, Anderson TA. Plant microRNA: a small regulatory molecule with big impact. Dev Biol. 2006, 289, 3-16.
52. Esmaeili F, Shiran B, Fallahi H, Mirakhorli N, Budak H, Martínez-Gómez P. In silico search and biological validation of microRNAs related to drought response in peach and almond. Funct Integr Genomics. 2017, 17, 189-201.
53. Ding Y, Tao Y, Zhu C. Emerging roles of microRNAs in the mediation of drought stress response in plants. J Exp Bot. 2013, 64, 3077-3086.
54. Chen H, Li Z, Xiong L. A plant microRNA regulates the adaptation of roots to drought stress. FEBS Letters. 2012, 586, 1742-1747.
55. Gao P, Bai X, Yang L, Lv D, Pan X, Li Y, Cai H, Ji W, Chen Q, Zhu Y. Osa-MIR393: a salinity- and alkaline stress-related microRNA gene. Mol Biol Rep. 2011, 38, 237-242.
56. Shi G-q, Fu J-y, Rong L-j, Zhang P-y, Guo C-j, Xiao K. TaMIR1119, a miRNA family member of wheat (Triticum aestivum), is essential in the regulation of plant drought tolerance. J Integr Agr. 2018, 17, 2369-2378.
57. Wang J, Ye Y, Xu M, Feng L, Xu LA. Roles of the SPL gene family and miR156 in the salt stress responses of tamarisk (Tamarix chinensis). BMC Plant Biol. 2019, 19, 370.
58. Giacomelli JI, Weigel D, Chan RL, Manavella PA. Role of recently evolved miRNA regulation of sunflower HaWRKY6 in response to temperature damage. New Phytol. 2012, 195, 766-773.
59. Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004, 16, 2001-2019.
60. Dong C, Pei H. Over-expression of miR397 improves plant tolerance to cold stress in Arabidopsis thaliana. J Plant Biol. 2014, 57, 209-217.
61. Thiebaut F, Rojas CA, Almeida KL, Grativol C, Domiciano GC, Lamb CR, Engler Jde A, Hemerly AS, Ferreira PC. Regulation of miR319 during cold stress in sugarcane. Plant Cell Environ. 2012, 35, 502-512.
62. Lv DK, Bai X, Li Y, Ding XD, Ge Y, Cai H, Ji W, Wu N, Zhu YM. Profiling of cold-stress-responsive miRNAs in rice by microarrays. Gene. 2010, 459, 39-47.
63. Wang ST, Sun XL, Hoshino Y, Yu Y, Jia B, Sun ZW, Sun MZ, Duan XB, Zhu YM. MicroRNA319 positively regulates cold tolerance by targeting OsPCF6 and OsTCP21 in rice (Oryza sativa L.). PLoS One. 2014, 9, e91357.
64. Valiollahi E, Farsi M, Kakhki AM. Sly-miR166 and Sly-miR319 are components of the cold stress response in Solanum lycopersicum. Plant Biotechnol Rep. 2014, 8, 349-356.
65. Lv DW, Zhen S, Zhu GR, Bian YW, Chen GX, Han CX, Yu ZT, Yan YM. High-throughput sequencing reveals H2O2 stress-associated microRNAs and a potential regulatory network in Brachypodium distachyon seedlings. Front Plant Sci. 2016, 7, 1567.
66. Bai Q, Wang X, Chen X, Shi G, Liu Z, Guo C, Xiao K. Wheat miRNA TaemiR408 acts as an essential mediator in plant tolerance to Pi deprivation and salt stress via modulating stress-associated physiological processes. Front Plant Sci. 2018, 9, 499.
67. Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD. Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci U S A. 2008, 105, 803-808.
68. Chauhan S, Yogindran S, Rajam MV. Role of miRNAs in biotic stress reactions in plants. Ind J Plant Physiol. 2017, 22, 514-529.
69. Zhu X, He S, Fang D, Guo L, Zhou X, Guo Y, Gao L, Qiao Y. High-throughput sequencing-based identification of Arabidopsis miRNAs induced by phytophthora capsici Infection. Front microbiol. 2020, 11, 1094.
70. Zhao X, Shan Y, Zhao Y, Wang A, Wang Z. A novel Arabidopsis miRNA, ath-miR38-3P, is involved in response to Sclerotinia sclerotiorum infection. J Integr Agr. 2016, 15, 2556-2562.
71. Xie S, Jiang H, Xu Z, Xu Q, Cheng B. Small RNA profiling reveals important roles for miRNAs in Arabidopsis response to Bacillus velezensis FZB42. Gene. 2017, 629, 9-15.
72. Xin M, Wang Y, Yao Y, Xie C, Peng H, Ni Z, Sun Q. Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol. 2010, 10, 123.
73. Feng J, Liu S, Wang M, Lang Q, Jin C. Identification of microRNAs and their targets in tomato infected with Cucumber mosaic virus based on deep sequencing. Planta. 2014, 240, 1335-1352.
74. Zhang X, Bao Y, Shan D, Wang Z, Song X, Wang Z, Wang J, He L, Wu L, Zhang Z, Niu D, Jin H, Zhao H. Magnaporthe oryzae induces the expression of a microRNA to suppress the immune response in rice. Plant Physiol. 2018, 177, 352-368.
75. Zhu QH, Fan L, Liu Y, Xu H, Llewellyn D, Wilson I. miR482 regulation of NBS-LRR defense genes during fungal pathogen infection in cotton. PLoS One. 2013, 8, e84390.
76. Yang L, Mu X, Liu C, Cai J, Shi K, Zhu W, Yang Q. Overexpression of potato miR482e enhanced plant sensitivity to Verticillium dahliae infection. J Integr Plant Biol. 2015, 57, 1078-1088.
77. Su Y, Li HG, Wang Y, Li S, Wang HL, Yu L, He F, Yang Y, Feng CH, Shuai P, Liu C, Yin W, Xia X. Poplar miR472a targeting NBS-LRRs is involved in effective defence against the necrotrophic fungus Cytospora chrysosperma. J Exp Bot. 2018, 69, 5519-5530.
78. Yan J, Zhang H, Zheng Y, Ding Y. Comparative expression profiling of miRNAs between the cytoplasmic male sterile line MeixiangA and its maintainer line MeixiangB during rice anther development. Planta. 2015, 241, 109-123.
79. Yu Y, Sun F, Chen N, Sun G, Wang CY, Wu DX. MiR396 regulatory network and its expression during grain development in wheat. Protoplasma. 2021, 258, 103-113.
80. Silva EM, Silva GFFe, Bidoia DB, Silva Azevedo M, Jesus FA, Pino LE, Peres LEP, Carrera E, López‐Díaz I, Nogueira FTS. MicroRNA159‐targeted SlGAMYB transcription factors are required for fruit set in tomato. Plant J. 2017, 92, 95-109.
81. Zhang Z, Wei L, Zou X, Tao Y, Liu Z, Zheng Y. Submergence-responsive microRNAs are potentially involved in the regulation of morphological and metabolic adaptations in maize root cells. Ann Bot. 2008, 102, 509-519.
82. Csukasi F, Donaire L, Casanal A, Martinez-Priego L, Botella MA, Medina-Escobar N, Llave C, Valpuesta V. Two strawberry miR159 family members display developmental-specific expression patterns in the fruit receptacle and cooperatively regulate Fa-GAMYB. New Phytol. 2012, 195, 47-57.
83. Ortiz-Morea FA, Vicentini R, Silva GF, Silva EM, Carrer H, Rodrigues AP, Nogueira FT. Global analysis of the sugarcane microtranscriptome reveals a unique composition of small RNAs associated with axillary bud outgrowth. J Exp Bot. 2013, 64, 2307-2320.
84. Xing L, Zhang D, Zhao C, Li Y, Ma J, An N, Han M. Shoot bending promotes flower bud formation by miRNA-mediated regulation in apple (Malus domestica Borkh.). Plant Biotechnol J. 2016, 14, 749-770.
85. Yang Z, Zhu P, Kang H, Liu L, Cao Q, Sun J, Dong T, Zhu M, Li Z, Xu T. High-throughput deep sequencing reveals the important role that microRNAs play in the salt response in sweet potato (Ipomoea batatas L.). BMC Genomics. 2020, 21, 164.
86. Li X, Bian H, Song D, Ma S, Han N, Wang J, Zhu M. Flowering time control in ornamental gloxinia (Sinningia speciosa) by manipulation of miR159 expression. Ann Bot. 2013, 111, 791-799.
87. Xu P, Zhu Y, Zhang Y, Jiang J, Yang L, Mu J, Yu X, He Y. Global analysis of the genetic variations in miRNA-targeted sites and their correlations with agronomic traits in rapeseed. Front Genet. 2021, 12, 741858.
88. Xu MY, Dong Y, Zhang QX, Zhang L, Luo YZ, Sun J, Fan YL, Wang L. Identification of miRNAs and their targets from Brassica napus by high-throughput sequencing and degradome analysis. BMC Genomics. 2012, 13, 421.
89. Xie FL, Huang SQ, Guo K, Xiang AL, Zhu YY, Nie L, Yang ZM. Computational identification of novel microRNAs and targets in Brassica napus. FEBS Lett. 2007, 581, 1464-1474.
90. Cheng H, Hao M, Wang W, Mei D, Tong C, Wang H, Liu J, Fu L, Hu Q. Genomic identification, characterization and differential expression analysis of SBP-box gene family in Brassica napus. BMC Plant Biol. 2016, 16, 196.
91. Cheng H, Hao M, Wang W, Mei D, Wells R, Liu J, Wang H, Sang S, Tang M, Zhou R, Chu W, Fu L, Hu Q. Integrative RNA- and miRNA-profile analysis reveals a likely role of BR and auxin signaling in branch angle regulation of B. napus. Int J Mol Sci. 2017, 18, 887.
92. Wang T, Ping X, Cao Y, Jian H, Gao Y, Wang J, Tan Y, Xu X, Lu K, Li J, Liu L. Genome-wide exploration and characterization of miR172/euAP2 genes in Brassica napus L. for likely role in flower organ development. BMC Plant Biol. 2019, 19, 336.
93. Jiang J, Xu P, Li Y, Li Y, Zhou X, Jiang M, Zhang J, Zhu J, Wang W, Yang L. Identification of miRNAs and their target genes in genic male sterility lines in Brassica napus by small RNA sequencing. BMC Plant Biol. 2021, 21, 520.
94. Korbes AP, Machado RD, Guzman F, Almerao MP, de Oliveira LF, Loss-Morais G, Turchetto-Zolet AC, Cagliari A, dos Santos Maraschin F, Margis-Pinheiro M, Margis R. Identifying conserved and novel microRNAs in developing seeds of Brassica napus using deep sequencing. PLoS One. 2012, 7, e50663.
95. Chen L, Chen L, Zhang X, Liu T, Niu S, Wen J, Yi B, Ma C, Tu J, Fu T, Shen J. Identification of miRNAs that regulate silique development in Brassica napus. Plant Sci. 2018, 269, 106-117.
96. Chen Z, Huo Q, Yang H, Jian H, Qu C, Lu K, Li J. Joint RNA-Seq and miRNA profiling analyses to reveal molecular mechanisms in regulating thickness of pod canopy in Brassica napus. Genes. 2019, 10, 591.
97. Huang D, Koh C, Feurtado JA, Tsang EW, Cutler AJ. MicroRNAs and their putative targets in Brassica napus seed maturation. BMC Genomics. 2013, 14, 140.
98. Wei W, Li G, Jiang X, Wang Y, Ma Z, Niu Z, Wang Z, Geng X. Small RNA and degradome profiling involved in seed development and oil synthesis of Brassica napus. PLoS One. 2018, 13, e0204998.
99. Tan M, Niu J, Peng DZ, Cheng Q, Luan MB, Zhang ZQ. Clone and function verification of the OPR gene in Brassica napus related to linoleic acid synthesis. BMC Plant Biol. 2022, 22, 192.
100. Zhao YT, Wang M, Fu SX, Yang WC, Qi CK, Wang XJ. Small RNA profiling in two Brassica napus cultivars identifies microRNAs with oil production- and development-correlated expression and new small RNA classes. Plant Physiol. 2012, 158, 813-823.
101. Wang J, Jian H, Wang T, Wei L, Li J, Li C, Liu L. Identification of microRNAs actively involved in fatty acid biosynthesis in developing Brassica napus seeds using high-throughput sequencing. Front Plant Sci. 2016, 7, 1570.
102. Wang Z, Qiao Y, Zhang J, Shi W, Zhang J. Genome wide identification of microRNAs involved in fatty acid and lipid metabolism of Brassica napus by small RNA and degradome sequencing. Gene. 2017, 619, 61-70.
103. Singroha G, Sharma P, Sunkur R. Current status of microRNA-mediated regulation of drought stress responses in cereals. Physiol Plant. 2021, 172, 1808-1821.
104. Cheng X, He Q, Tang S, Wang H, Zhang X, Lv M, Liu H, Gao Q, Zhou Y, Wang Q, Man X, Liu J, Huang R, Wang H, Chen T, Liu J. The miR172/IDS1 signaling module confers salt tolerance through maintaining ROS homeostasis in cereal crops. New Phytol. 2021, 230, 1017-1033.
105. Lantzouni O, Alkofer A, Falter-Braun P, Schwechheimer C. Growth-regulating factors interact with DELLAs and regulate growth in cold stress. Plant Cell. 2020, 32, 1018-1034.
106. Pegler JL, Oultram JMJ, Nguyen DQ, Grof CPL, Eamens AL. MicroRNA-mediated responses to cadmium stress in Arabidopsis thaliana. Plants. 2021, 10, 130.
107. Eshkiki EM, Hajiahmadi Z, Abedi A, Kordrostami M, Jacquard C. In Silico analyses of autophagy-related genes in rapeseed (Brassica napus L.) under different abiotic stresses and in various tissues. Plants. 2020, 9, 1393.
108. Jatan R, Lata C. Role of microRNAs in abiotic and biotic stress resistance in plants. P Indian Natl Sci Ac. 2019, 85, 553-567.
109. Jian H, Wang J, Wang T, Wei L, Li J, Liu L. Identification of rapeseed microRNAs involved in early stage seed germination under salt and drought stresses. Front Plant Sci. 2016, 7, 658.
110. Li J, Duan Y, Sun N, Wang L, Feng S, Fang Y, Wang Y. The miR169n-NF-YA8 regulation module involved in drought resistance in Brassica napus L. Plant Sci. 2021, 313, 111062.
111. Megha S, Basu U, Joshi RK, Kav NNV. Physiological studies and genome-wide microRNA profiling of cold-stressed Brassica napus. Plant Physiol Biochem. 2018, 132, 1-17.
112. Huang SQ, Xiang AL, Che LL, Chen S, Li H, Song JB, Yang ZM. A set of miRNAs from Brassica napus in response to sulphate deficiency and cadmium stress. Plant Biotechnol J. 2010, 8, 887-899.
113. Jian H, Yang B, Zhang A, Ma J, Ding Y, Chen Z, Li J, Xu X, Liu L. Genome-wide identification of microRNAs in response to cadmium stress in oilseed rape (Brassica napus L.) using high-throughput sequencing. Int J Mol Sci. 2018, 19, 1431.
114. Fu Y, Mason AS, Zhang Y, Lin B, Xiao M, Fu D, Yu H. MicroRNA-mRNA expression profiles and their potential role in cadmium stress response in Brassica napus. BMC Plant Biol. 2019, 19, 570.
115. Zhou ZS, Song JB, Yang ZM. Genome-wide identification of Brassica napus microRNAs and their targets in response to cadmium. J Exp Bot. 2012, 63, 4597-4613.
116. Meng JG, Zhang XD, Tan SK, Zhao KX, Yang ZM. Genome-wide identification of Cd-responsive NRAMP transporter genes and analyzing expression of NRAMP 1 mediated by miR167 in Brassica napus. Biometals. 2017, 30, 917-931.
117. Zhang LW, Song JB, Shu XX, Zhang Y, Yang ZM. miR395 is involved in detoxification of cadmium in Brassica napus. J Hazard Mater. 2013, 250-251, 204-211.
118. Zhang XD, Sun JY, You YY, Song JB, Yang ZM. Identification of Cd-responsive RNA helicase genes and expression of a putative BnRH 24 mediated by miR158 in canola (Brassica napus). Ecotoxicol Environ Saf. 2018, 157, 159-168.
119. He X, Zhang H, Ye X, Hong J, Ding G. Nitrogen assimilation related genes in Brassica napus: systematic characterization and expression analysis identified hub genes in multiple nutrient stress responses. Plants. 2021, 10, 2160.
120. Buhtz A, Springer F, Chappell L, Baulcombe DC, Kehr J. Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J. 2008, 53, 739-749.
121. Pant BD, Buhtz A, Kehr J, Scheible WR. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 2008, 53, 731-738.
122. Buhtz A, Pieritz J, Springer F, Kehr J. Phloem small RNAs, nutrient stress responses, and systemic mobility. BMC Plant Biol. 2010, 10, 64.
123. Pant BD, Musialak-Lange M, Nuc P, May P, Buhtz A, Kehr J, Walther D, Scheible WR. Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol. 2009, 150, 1541-1555.
124. Hua YP, Zhou T, Huang JY, Yue CP, Song HX, Guan CY, Zhang ZH. Genome-wide differential DNA methylation and miRNA expression profiling reveals epigenetic regulatory mechanisms underlying nitrogen-limitation-triggered adaptation and use efficiency enhancement in allotetraploid rapeseed. Int J Mol Sci. 2020, 21, 8453.
125. Shen D, Suhrkamp I, Wang Y, Liu S, Menkhaus J, Verreet JA, Fan L, Cai D. Identification and characterization of microRNAs in oilseed rape (Brassica napus) responsive to infection with the pathogenic fungus Verticillium longisporum using Brassica AA (Brassica rapa) and CC (Brassica oleracea) as reference genomes. New Phytol. 2014, 204, 577-594.
126. Cao JY, Xu YP, Zhao L, Li SS, Cai XZ. Tight regulation of the interaction between Brassica napus and Sclerotinia sclerotiorum at the microRNA level. Plant Mol Biol. 2016, 92, 39-55.
127. Jian H, Ma J, Wei L, Liu P, Zhang A, Yang B, Li J, Xu X, Liu L. Integrated mRNA, sRNA, and degradome sequencing reveal oilseed rape complex responses to Sclerotinia sclerotiorum (Lib.) infection. Sci Rep. 2018, 8, 10987.
128. Regmi R, Newman TE, Kamphuis LG, Derbyshire MC. Identification of B. napus small RNAs responsive to infection by a necrotrophic pathogen. BMC Plant Biol. 2021, 21, 366.
129. Verma SS, Rahman MH, Deyholos MK, Basu U, Kav NN. Differential expression of miRNAs in Brassica napus root following infection with Plasmodiophora brassicae. PLoS One. 2014, 9, e86648.
130. Depotter JRL, Deketelaere S, Inderbitzin P, Tiedemann AV, Höfte M, Subbarao KV, Wood TA, Thomma BPHJ. Verticillium longisporum, the invisible threat to oilseed rape and other brassicaceous plant hosts. Mol Plant Pathol. 2016, 17, 1004-1016.
131. Jian WA, Sy A, Li LA, Dl A, Sr A, Wz A, Wm A, Pc A, Qs B, Yf B. Host-induced gene silencing of multiple pathogenic factors of Sclerotinia sclerotiorum confers resistance to sclerotinia rot in Brassica napus. Crop J. 2021, 10, 661-671.
132. Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006, 312, 436-439.
133. Chalhoub B, Denoeud F, Liu S, Parkin IAP, Tang H, Wang X, Chiquet J, Belcram H, Tong C, Samans B, Corréa M, Da Silva C, Just J, Falentin C, Koh CS, Le Clainche I, Bernard M, Bento P, Noel B, Labadie K, Alberti A, Charles M, Arnaud D, Guo H, Daviaud C, Alamery S, Jabbari K, Zhao M, Edger PP, Chelaifa H, Tack D, Lassalle G, Mestiri I, Schnel N, Le Paslier M-C, Fan G, Renault V, Bayer PE, Golicz AA, Manoli S, Lee T-H, Thi VHD, Chalabi S, Hu Q, Fan C, Tollenaere R, Lu Y, Battail C, Shen J, Sidebottom CHD, Wang X, Canaguier A, Chauveau A, Bérard A, Deniot G, Guan M, Liu Z, Sun F, Lim YP, Lyons E, Town CD, Bancroft I, Wang X, Meng J, Ma J, Pires JC, King GJ, Brunel D, Delourme R, Renard M, Aury J-M, Adams KL, Batley J, Snowdon RJ, Tost J, Edwards D, Zhou Y, Hua W, Sharpe AG, Paterson AH, Guan C, Wincker P. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014, 345, 950-953.
134. Dai X, Zhuang Z, Zhao PX. psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, 49-54.
135. Chen C, Liu C, Jiang A, Zhao Q, Zhang Y, Hu W. miRNA and degradome sequencing identify miRNAs and their target genes involved in the browning inhibition of fresh-cut apples by hydrogen sulfide. J Agric Food Chem. 2020, 68, 8462-8470.
136. Kunert KJ, Vorster BJ, Fenta BA, Kibido T, Dionisio G, Foyer CH. Drought stress responses in soybean roots and nodules. Front Plant Sci. 2016, 7, 1015.
137. Zilli JÉ, Pacheco RS, Gianluppi V, Smiderle OJ, Urquiaga S, Hungria M. Biological N2 fixation and yield performance of soybean inoculated with Bradyrhizobium. Nutr Cycl Agroecosys. 2021, 119, 323-336.
138. Ngalimat MS, Mohd Hata E, Zulperi D, Ismail SI, Ismail MR, Mohd Zainudin NAI, Saidi NB, Yusof MT. Plant growth-promoting bacteria as an emerging tool to manage bacterial rice pathogens. Microorganisms. 2021, 9, 682.
139. Mamet SD, Helgason BL, Lamb EG, McGillivray A, Stanley KG, Robinson SJ, Aziz SU, Vail S, Siciliano SD. Phenology-dependent root bacteria enhance yield of Brassica napus. Soil Biol Biochem. 2022, 166, 108468.
140. Dai C, Li Y, Li L, Du Z, Lin S, Tian X, Li S, Yang B, Yao W, Wang J, Guo L, Lu S. An efficient agrobacterium-mediated transformation method using hypocotyl as explants for Brassica napus. Mol Breeding. 2020, 40, 96.
141. Li F, Wang W, Zhao N, Xiao B, Cao P, Wu X, Ye C, Shen E, Qiu J, Zhu QH, Xie J, Zhou X, Fan L. Regulation of nicotine biosynthesis by an endogenous target mimicry of microRNA in tobacco. Plant Physiol. 2015, 169, 1062-1071.
142. Zhang H, Zhang J, Yan J, Gou F, Mao Y, Tang G, Botella JR, Zhu JK. Short tandem target mimic rice lines uncover functions of miRNAs in regulating important agronomic traits. Proc Natl Acad Sci U S A. 2017, 114, 5277-5282.
143. Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007, 39, 1033-1037.
144. Tang G, Yan J, Gu Y, Qiao M, Fan R, Mao Y, Tang X. Construction of short tandem target mimic (STTM) to block the functions of plant and animal microRNAs. Methods. 2012, 58, 118-125.
145. Gupta SK, Vishwakarma A, Kenea HD, Galsurker O, Cohen H, Aharoni A, Arazi T. CRISPR/Cas9 mutants of tomato microRNA164 genes uncover their functional specialization in development. Plant Physiol. 2021, 187, 1636-1652.
146. Lin Y, Zhu Y, Cui Y, Chen R, Chen Z, Li G, Fan M, Chen J, Li Y, Guo X, Zheng X, Chen L, Wang F. Derepression of specific miRNA-target genes in rice using CRISPR/Cas9. J Exp Bot. 2021, 72, 7067-7077.
147. Zheng M, Zhang L, Tang M, Liu J, Liu H, Yang H, Fan S, Terzaghi W, Wang H, Hua W. Knockout of two BnaMAX1 homologs by CRISPR/Cas9-targeted mutagenesis improves plant architecture and increases yield in rapeseed (Brassica napus L.). Plant Biotechnol J. 2020, 18, 644-654.
148. Fan S, Zhang L, Tang M, Cai Y, Liu J, Liu H, Liu J, Terzaghi W, Wang H, Hua W, Zheng M. CRISPR/Cas9-targeted mutagenesis of the BnaA03.BP gene confers semi-dwarf and compact architecture to rapeseed (Brassica napus L.). Plant Biotechnol J. 2021, 19, 2383-2385.
149. Zaman QU, Wen C, Yuqin S, Mengyu H, Desheng M, Jacqueline B, Baohong Z, Chao L, Qiong H. Characterization of SHATTERPROOF homoeologs and CRISPR-Cas9-mediated genome editing enhances pod-shattering resistance in Brassica napus L. Crispr j. 2021, 4, 360-370.
150. Zaman QU, Chu W, Hao M, Shi Y, Sun M, Sang SF, Mei D, Cheng H, Liu J, Li C, Hu Q. CRISPR/Cas9-mediated multiplex genome editing of JAGGED Gene in Brassica napus L. Biomolecules. 2019, 9, 725.
151. Khan MHU, Hu L, Zhu M, Zhai Y, Khan SU, Ahmar S, Amoo O, Zhang K, Fan C, Zhou Y. Targeted mutagenesis of EOD3 gene in Brassica napus L. regulates seed production. J Cell Physiol. 2021, 236, 1996-2007.
152. Xie T, Chen X, Guo T, Rong H, Chen Z, Sun Q, Batley J, Jiang J, Wang Y. Targeted knockout of BnTT2 Homologues for yellow-seeded Brassica napus with reduced flavonoids and improved fatty acid composition. J Agric Food Chem. 2020, 68, 5676-5690.
153. Raza A, Razzaq A, Mehmood SS, Hussain MA, Wei S, He H, Zaman QU, Xuekun Z, Hasanuzzaman M. Omics: The way forward to enhance abiotic stress tolerance in Brassica napus L. GM Crops Food. 2021, 12, 251-281.
This entry is adapted from the peer-reviewed paper 10.3390/life12111811