Antibodies (Abs) are important immune mediators and powerful diagnostic markers in a wide range of infectious diseases. Understanding the humoral immunity or the development of effective antibodies against SARS-CoV-2 is a prerequisite for limiting disease burden in the community and aids in the development of new diagnostic, therapeutic, and vaccination options. Antibody testing showed the potential in adding important diagnostic value to the routine diagnosis and clinical management of COVID-19. They could also play a critical role in COVID-19 surveillance, allowing for a better understanding of the full scope of the disease. The development of several vaccines and the success of passive immunotherapy suggest that anti-SARS-CoV-2 antibodies have the potential to be used in the treatment and prevention of SARS-CoV-2 infection.
- COVID-19
- SARS-CoV-2
- antibodies
- testing
- serodiagnostic
- immunotherapy
- vaccine
1. Introduction
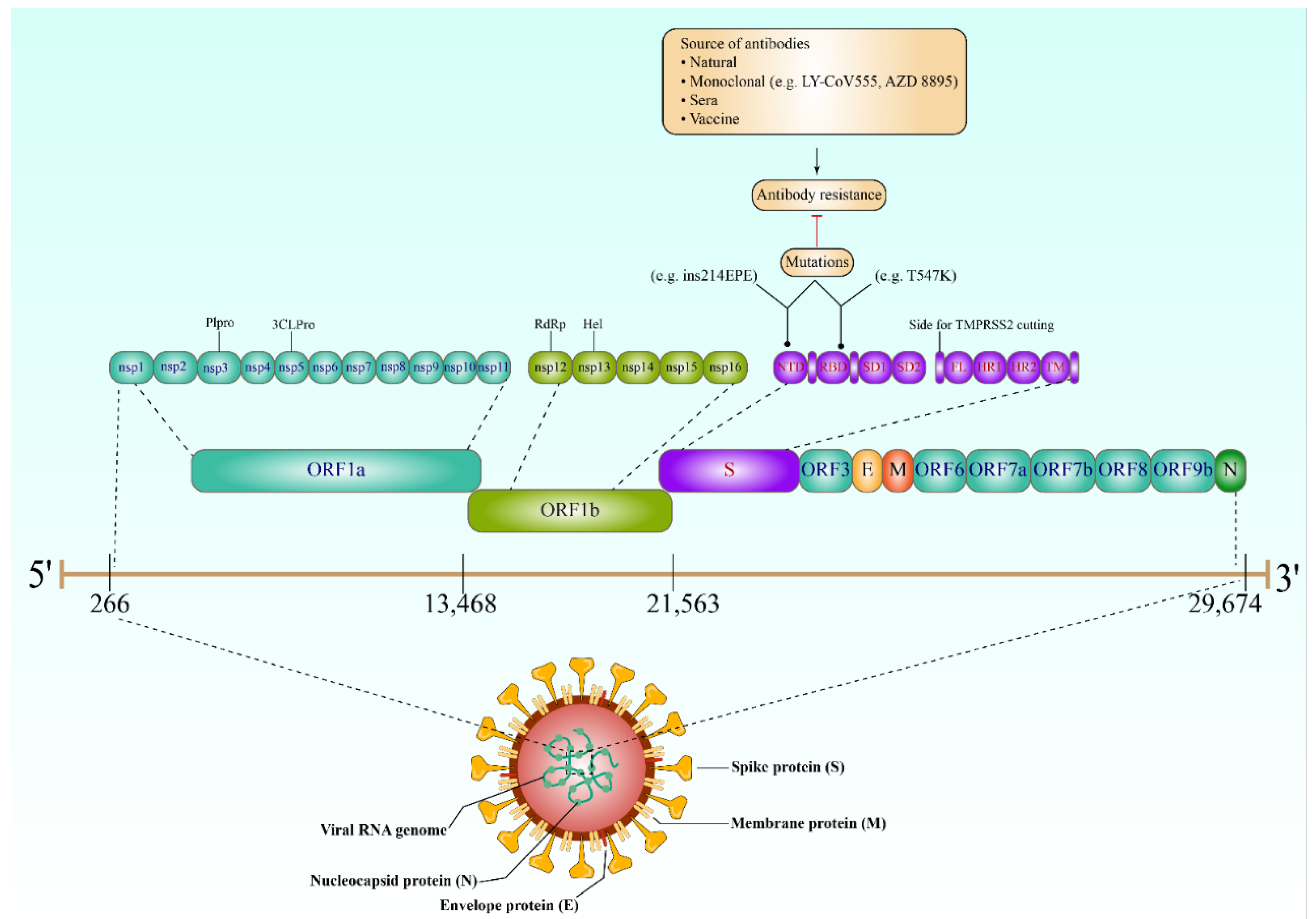
2. Key Proteins of SARS-CoV-2
3. Antibody Response against SARS-CoV-2
4. The Role of Antibodies in SARS-CoV-2 Diagnosis
4.1. COVID-19 Antibody Tests
4.2. Implications of Seroconversion in SARS-CoV-2 Antibody Tests
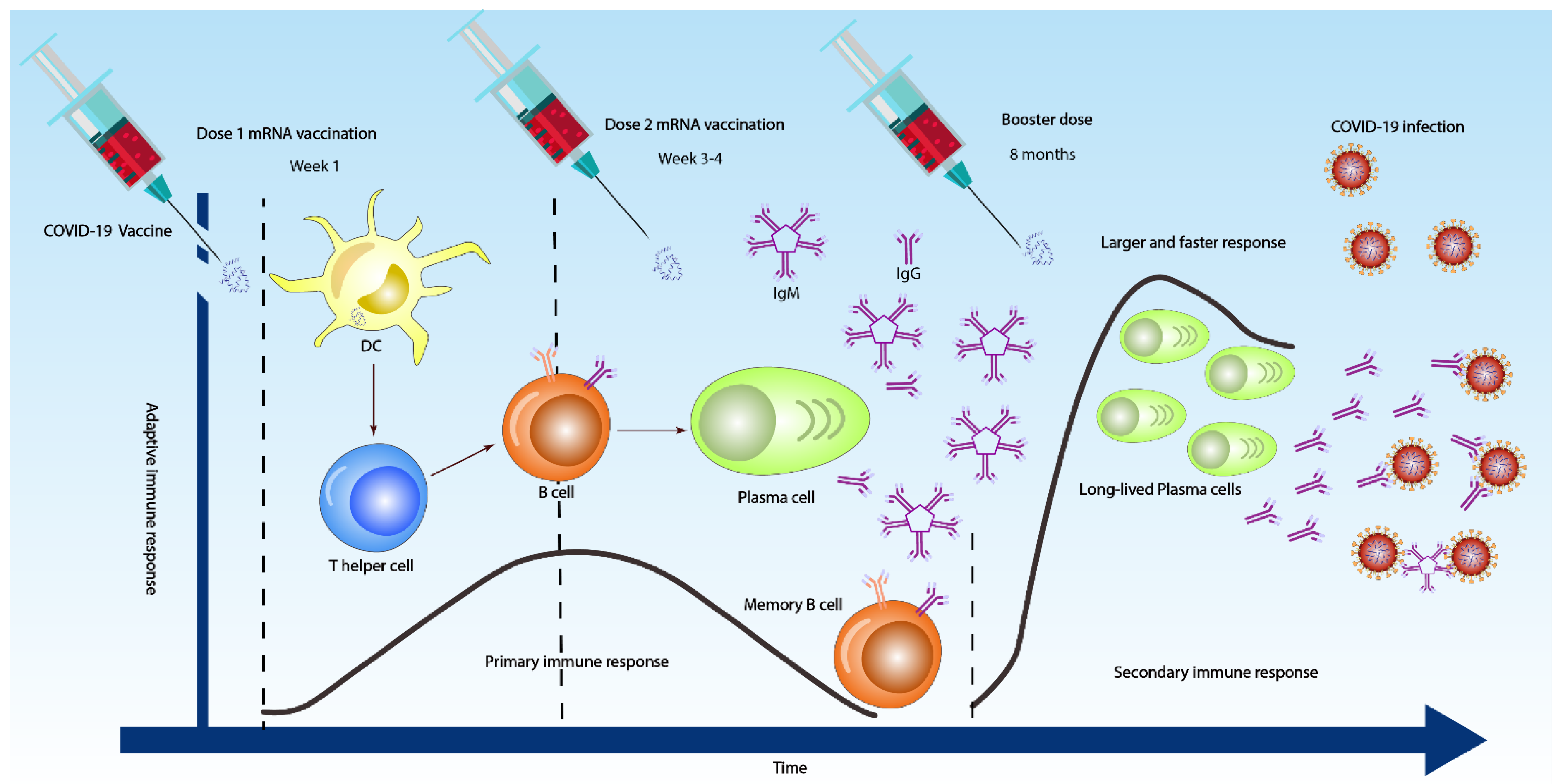
4.3. Antibody Tests at an Individual Level
4.4. Population Serological Testing
Serological testing can help identify who is infected or exposed and who is immune by assuming protective immunity. Accordingly, population-based serological information will be helpful for officials in making decisions about lifting or enforcing any control measures. The potential of this test in determining the accurate number of infected people in a large population has been tested [37][38]. Population seropositivity indicates that the number of people positive for anti-SARS-CoV-2 antibodies is much higher than that of the reported cases [38][39]. Population-based serological surveillance has been carried out around the world, including countries in Europe [40][41], America [42][43], Asia [44][45], and Africa [46][47]. Indeed, healthcare workers are the population most targeted for serological surveillance due to their high risk of SARS-CoV-2 infection, with ELISA being the most commonly used diagnostic tool for detecting anti-SARS-CoV-2 antibodies.
5. Antibody Tests and Seroprotection
5.1. Antibodies in SARS-CoV-2 Vaccine Development
5.2. Mechanism of Antibody-Mediated Protection
5.3. Prevaccination Antibody Screening
6. Resistance to SARS-CoV-2 Antibodies
7. Limitations of Antibody Tests
This entry is adapted from the peer-reviewed paper 10.3390/vaccines10081346
References
- Chowdhury, M.A.; Hossain, N.; Kashem, M.A.; Shahid, M.A.; Alam, A. Immune response in COVID-19: A review. J. Infect. Public Health 2020, 13, 1619–1629.
- Scourfield, D.O.; Reed, S.G.; Quastel, M.; Alderson, J.; Bart, V.M.; Teijeira Crespo, A.; Jones, R.; Pring, E.; Richter, F.C.; The Oxford-Cardiff COVID-19 Literature Consortium; et al. The role and uses of antibodies in COVID-19 infections: A living review. Oxf. Open Immunol. 2021, 2, iqab003.
- Mallano, A.; Ascione, A.; Flego, M. Antibody Response against SARS-CoV-2 Infection: Implications for Diagnosis, Treatment and Vaccine Development. Int. Rev. Immunol. 2021, 41, 393–413.
- Khailany, R.A.; Safdar, M.; Ozaslan, M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020, 19, 100682.
- Ching, L.; Chang, S.P.; Nerurkar, V.R. COVID-19 Special Column: Principles Behind the Technology for Detecting SARS-CoV-2, the Cause of COVID-19. Hawaii J. Health Soc. Welf. 2020, 79, 136.
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.I. Insights into SARS-CoV-2 Genome, Structure, Evolution, Pathogenesis and Therapies: Structural Genomics Approach. Biochim. Biophys. Acta Mol Basis Dis. 2020, 1, 165878.
- Chang, C.-k.; Sue, S.-C.; Yu, T.-h.; Hsieh, C.-M.; Tsai, C.-K.; Chiang, Y.-C.; Lee, S.-j.; Hsiao, H.-h.; Wu, W.-J.; Chang, W.-L. Modular organization of SARS coronavirus nucleocapsid protein. J. Biomed. Sci. 2006, 13, 59–72.
- Dispinseri, S.; Secchi, M.; Pirillo, M.F.; Tolazzi, M.; Borghi, M.; Brigatti, C.; De Angelis, M.L.; Baratella, M.; Bazzigaluppi, E.; Venturi, G.; et al. Neutralizing antibody responses to SARS-CoV-2 in symptomatic COVID-19 is persistent and critical for survival. Nat. Commun. 2021, 12, 2670.
- Amanat, F.; Stadlbauer, D.; Strohmeier, S.; Nguyen, T.H.; Chromikova, V.; McMahon, M.; Jiang, K.; Arunkumar, G.A.; Jurczyszak, D.; Polanco, J. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020, 26, 1033–1036.
- Carter, L.J.; Garner, L.V.; Smoot, J.W.; Li, Y.; Zhou, Q.; Saveson, C.J.; Sasso, J.M.; Gregg, A.C.; Soares, D.J.; Beskid, T.R. Assay techniques and test development for COVID-19 diagnosis. ACS Cent. Sci. 2020, 6, 591–605.
- Scohy, A.; Anantharajah, A.; Bodéus, M.; Kabamba-Mukadi, B.; Verroken, A.; Rodriguez-Villalobos, H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J. Clin. Virol. 2020, 129, 104455.
- To, K.K.-W.; Tsang, O.T.-Y.; Leung, W.-S.; Tam, A.R.; Wu, T.-C.; Lung, D.C.; Yip, C.C.-Y.; Cai, J.-P.; Chan, J.M.-C.; Chik, T.S.-H. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study. Lancet Infect. Dis. 2020, 20, 565–574.
- Udugama, B.; Kadhiresan, P.; Kozlowski, H.N.; Malekjahani, A.; Osborne, M.; Li, V.Y.; Chen, H.; Mubareka, S.; Gubbay, J.B.; Chan, W.C. Diagnosing COVID-19: The disease and tools for detection. ACS Nano 2020, 14, 3822–3835.
- Ma, H.; Zeng, W.; He, H. Serum IgA, IgM, and IgG Responses in COVID-19. Cell Mol Immunol. 2020, 17, 773–775.
- Cai, X.-F.; Chen, J.; Long, Q.-X.; Deng, H.-J.; Liu, P.; Fan, K.; Liao, P.; Liu, B.-Z.; Wu, G.-C.; Chen, Y.-K. A Peptide-Based Magnetic Chemiluminescence Enzyme Immunoassay for Serological Diagnosis of Coronavirus Disease 2019. J. Infect. Dis. 2020, 222, 189–193.
- Infantino, M.; Grossi, V.; Lari, B.; Bambi, R.; Perri, A.; Manneschi, M.; Terenzi, G.; Liotti, I.; Ciotta, G.; Taddei, C. Diagnostic accuracy of an automated chemiluminescent immunoassay for anti-SARS-CoV-2 IgM and IgG antibodies: An Italian experience. J. Med. Virol. 2020, 92, 1671–1675.
- Guo, L.; Ren, L.; Yang, S.; Xiao, M.; Chang, D.; Yang, F.; Dela Cruz, C.S.; Wang, Y.; Wu, C.; Xiao, Y. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin. Infect. Dis. 2020, 71, 778–785.
- Liu, W.; Liu, L.; Kou, G.; Zheng, Y.; Ding, Y.; Ni, W.; Wang, Q.; Tan, L.; Wu, W.; Tang, S. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020, 58, e00461-20.
- Chen, Z.; Zhang, Z.; Zhai, X.; Li, Y.; Lin, L.; Zhao, H.; Bian, L.; Li, P.; Yu, L.; Wu, Y. Rapid and Sensitive Detection of anti-SARS-CoV-2 IgG, Using Lanthanide-Doped Nanoparticles-Based Lateral Flow Immunoassay. Anal. Chem. 2020, 92, 7226–7231.
- Stephen, M.; Hahn, M.D.; Commissioner of Food and Drugs—Food and Drug Administration. Coronavirus (COVID-19) Update: FDA Authorizes First Antigen Test to Help in the Rapid Detection of the Virus that Causes COVID-19 in Patients. Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-first-antigen-test-help-rapid-detection-virus-causes (accessed on 26 July 2022).
- Zhao, J.; Yuan, Q.; Wang, H.; Liu, W.; Liao, X.; Su, Y.; Wang, X.; Yuan, J.; Li, T.; Li, J. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020, 71, 2027–2034.
- Fu, Y.; Li, Y.; Guo, E.; He, L.; Liu, J.; Yang, B.; Li, F.; Wang, Z.; Li, Y.; Xiao, R.; et al. SARS-CoV-2 Viral Dynamic Shedding, Seroconversion, and Diseases Severity Among 3192 Patients Hospitalized with COVID-19: A Large Case Series Study in Wuhan. Lancet 2020. Available online: https://ssrn.com/abstract=3594600 or http://dx.doi.org/10.2139/ssrn.3594600 (accessed on 2 July 2022).
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469.
- Wu, L.-P.; Wang, N.-C.; Chang, Y.-H.; Tian, X.-Y.; Na, D.-Y.; Zhang, L.-Y.; Zheng, L.; Lan, T.; Wang, L.-F.; Liang, G.-D. Duration of antibody responses after severe acute respiratory syndrome. Emerg. Infect. Dis. 2007, 13, 1562.
- Zhang, W.; Du, R.-H.; Li, B.; Zheng, X.-S.; Yang, X.-L.; Hu, B.; Wang, Y.-Y.; Xiao, G.-F.; Yan, B.; Shi, Z.-L. Molecular and serological investigation of 2019-nCoV infected patients: Implication of multiple shedding routes. Emerg. Microbes Infect. 2020, 9, 386–389.
- Pan, Y.; Li, X.; Yang, G.; Fan, J.; Tang, Y.; Zhao, J.; Long, X.; Guo, S.; Zhao, Z.; Liu, Y. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. J. Infect. 2020, 81, e28–e32.
- West, R.; Kobokovich, A.; Connell, N.; Gronvall, G.K. COVID-19 Antibody Tests: A Valuable Public Health Tool with Limited Relevance to Individuals. Trends Microbiol. 2020, 29, 214–223.
- Hoffman, T.; Nissen, K.; Krambrich, J.; Rönnberg, B.; Akaberi, D.; Esmaeilzadeh, M.; Salaneck, E.; Lindahl, J.; Lundkvist, Å. Evaluation of a COVID-19 IgM and IgG rapid test; an efficient tool for assessment of past exposure to SARS-CoV-2. Infect. Ecol. Epidemiol. 2020, 10, 1754538.
- Xiang, F.; Wang, X.; He, X.; Peng, Z.; Yang, B.; Zhang, J.; Zhou, Q.; Ye, H.; Ma, Y.; Li, H. Antibody detection and dynamic characteristics in patients with COVID-19. Clin. Infect. Dis. 2020, 71, 1930–1934.
- CDC. Interim Guidelines for COVID-19 Antibody. Testing: What are the implications? Available online: https://stacks.cdc.gov/view/cdc/91257 (accessed on 1 July 2022).
- Abbasi, J. The promise and peril of antibody testing for COVID-19. JAMA 2020, 323, 1881–1883.
- Cassaniti, I.; Novazzi, F.; Giardina, F.; Salinaro, F.; Sachs, M.; Perlini, S.; Bruno, R.; Mojoli, F.; Baldanti, F. Performance of VivaDiag COVID-19 IgM/IgG Rapid Test is inadequate for diagnosis of COVID-19 in acute patients referring to emergency room department. J. Med. Virol. 2020, 92, 1724–1727.
- Li, Z.; Yi, Y.; Luo, X.; Xiong, N.; Liu, Y.; Li, S.; Sun, R.; Wang, Y.; Hu, B.; Chen, W. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020, 92, 1724–1727.
- WHO. Advice on the Use of Point-of-Care Immunodiagnostic Tests for COVID-19; Scientific Brief. 2020. Available online: https://www.who.int/news-room/commentaries/detail/advice-on-the-use-of-point-of-care-immunodiagnostic-tests-for-covid-19 (accessed on 26 July 2022).
- Lv, H.; Wu, N.C.; Tsang, O.T.-Y.; Yuan, M.; Perera, R.A.; Leung, W.S.; So, R.T.; Chan, J.M.C.; Yip, G.K.; Chik, T.S.H. Cross-reactive antibody response between SARS-CoV-2 and SARS-CoV infections. Cell Rep. 2020, 31, 107725.
- Jacofsky, D.; Jacofsky, E.M.; Jacofsky, M. Understanding antibody testing for COVID-19. J. Arthroplast. 2020, 35, S74–S81.
- Bendavid, E.; Mulaney, B.; Sood, N.; Shah, S.; Ling, E.; Bromley-Dulfano, R.; Lai, C.; Weissberg, Z.; Saavedra, R.; Tedrow, J. COVID-19 Antibody Seroprevalence in Santa Clara County, California. Int. J. Epidemiol. 2021, 50, 410–419.
- Doi, A.; Iwata, K.; Kuroda, H.; Hasuike, T.; Nasu, S.; Kanda, A.; Nagao, T.; Nishioka, H.; Tomii, K.; Morimoto, T. Estimation of seroprevalence of novel coronavirus disease (COVID-19) using preserved serum at an outpatient setting in Kobe, Japan: A cross-sectional study. Clin. Epidemiology Glob. Heal. 2021, 11, 100747.
- Shakiba, M.; Nazari, S.S.H.; Mehrabian, F.; Rezvani, S.M.; Ghasempour, Z.; Heidarzadeh, A. Seroprevalence of COVID-19 virus infection in Guilan province, Iran. Emerg. Infect Dis. 2020, 27, 636–638.
- Pollán, M.; Pérez-Gómez, B.; Pastor-Barriuso, R.; Oteo, J.; Hernán, M.A.; Pérez-Olmeda, M.; Sanmartín, J.L.; Fernández-García, A.; Cruz, I.; de Larrea, N.F. Prevalence of SARS-CoV-2 in Spain (ENE-COVID): A nationwide, population-based seroepidemiological study. Lancet 2020, 396, 535–544.
- Stringhini, S.; Wisniak, A.; Piumatti, G.; Azman, A.S.; Lauer, S.A.; Baysson, H.; De Ridder, D.; Petrovic, D.; Schrempft, S.; Marcus, K. Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Geneva, Switzerland (SEROCoV-POP): A population-based study. Lancet 2020, 396, 313–319.
- Silveira, M.F.; Barros, A.J.; Horta, B.L.; Pellanda, L.C.; Victora, G.D.; Dellagostin, O.A.; Struchiner, C.J.; Burattini, M.N.; Valim, A.R.; Berlezi, E.M. Population-based surveys of antibodies against SARS-CoV-2 in Southern Brazil. Nat. Med. 2020, 26, 1196–1199.
- Havers, F.P.; Reed, C.; Lim, T.; Montgomery, J.M.; Klena, J.D.; Hall, A.J.; Fry, A.M.; Cannon, D.L.; Chiang, C.-F.; Gibbons, A.; et al. Seroprevalence of antibodies to SARS-CoV-2 in 10 sites in the United States, 23 March–12 May 2020. JAMA Intern. Med. 2020, 180, 1576–1586.
- Xu, X.; Sun, J.; Nie, S.; Li, H.; Kong, Y.; Liang, M.; Hou, J.; Huang, X.; Li, D.; Ma, T. Seroprevalence of immunoglobulin M and G antibodies against SARS-CoV-2 in China. Nat. Med. 2020, 26, 1193–1195.
- Noh, J.Y.; Seo, Y.B.; Yoon, J.G.; Seong, H.; Hyun, H.; Lee, J.; Lee, N.; Jung, S.; Park, M.-J.; Song, W. Seroprevalence of anti-SARS-CoV-2 antibodies among outpatients in southwestern Seoul, Korea. J. Korean Med. Sci. 2020, 35, e311.
- Adetifa, I.M.; Uyoga, S.; Gitonga, J.N.; Mugo, D.; Otiende, M.; Nyagwange, J.; Karanja, H.K.; Tuju, J.; Wanjiku, P.; Aman, R. Temporal trends of SARS-CoV-2 seroprevalence in transfusion blood donors during the first wave of the COVID-19 epidemic in Kenya. Nat. Commun. 2021, 12, 3966.
- George, J.A.; Khoza, S.; Mayne, E.; Dlamini, S.; Kone, N.; Jassat, W.; Chetty, K.; Centner, C.; Pillay, T.; Maphayi, M. Sentinel seroprevalence of SARS-CoV-2 in Gauteng Province, South Africa, August–October 2020. S. Afr. Med. J. 2021, 111, 1078–1083.
- Peng, R.; Wu, L.-A.; Wang, Q.; Qi, J.; Gao, G.F. Cell entry by SARS-CoV-2. Trends Biochem. Sci. 2021, 46, 848–860.
- Alzain, A.A.; Elbadwi, F.A.; Alsamani, F.O. Discovery of novel TMPRSS2 inhibitors for COVID-19 using in silico fragment-based drug design, molecular docking, molecular dynamics, and quantum mechanics studies. Inform. Med. Unlocked 2022, 29, 100870.
- Lo Sasso, B.; Agnello, L.; Giglio, R.V.; Gambino, C.M.; Ciaccio, A.M.; Vidali, M.; Ciaccio, M. Longitudinal analysis of anti-SARS-CoV-2 S-RBD IgG antibodies before and after the third dose of the BNT162b2 vaccine. Sci. Rep. 2022, 12, 8679.
- Huang, A.T.; Garcia-Carreras, B.; Hitchings, M.D.T.; Yang, B.; Katzelnick, L.C.; Rattigan, S.M.; Borgert, B.A.; Moreno, C.A.; Solomon, B.D.; Trimmer-Smith, L.; et al. A systematic review of antibody mediated immunity to coronaviruses: Kinetics, correlates of protection, and association with severity. Nat. Commun. 2020, 11, 4704.
- Park, Y.-J.; Marco, A.D.; Starr, T.N.; Liu, Z.; Pinto, D.; Walls, A.C.; Zatta, F.; Zepeda, S.K.; Bowen, J.E.; Sprouse, K.R.; et al. Antibody-mediated broad sarbecovirus neutralization through ACE2 molecular mimicry. Science 2022, 375, 449–454.
- Hansen, J.; Baum, A.; Pascal, K.E.; Russo, V.; Giordano, S.; Wloga, E.; Fulton, B.O.; Yan, Y.; Koon, K.; Patel, K. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 2020, 369, 1010–1014.
- Shi, R.; Shan, C.; Duan, X.; Chen, Z.; Liu, P.; Song, J.; Song, T.; Bi, X.; Han, C.; Wu, L. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 2020, 584, 120–124.
- Frieman, M.; Harris, A.D.; Herati, R.S.; Krammer, F.; Mantovani, A.; Rescigno, M.; Sajadi, M.M.; Simon, V. SARS-CoV-2 vaccines for all but a single dose for COVID-19 survivors. EBioMedicine 2021, 68, 103401.
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424.
- Faria, N.R.; Mellan, T.A.; Whittaker, C.; Claro, I.M.; Candido, D.d.S.; Mishra, S.; Crispim, M.A.E.; Sales, F.C.S.; Hawryluk, I.; McCrone, J.T.; et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 2021, 372, 815–821.
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 2021, 593, 130–135.
- Li, Q.; Nie, J.; Wu, J.; Zhang, L.; Ding, R.; Wang, H.; Zhang, Y.; Li, T.; Liu, S.; Zhang, M.; et al. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell 2021, 184, 2362–2371.e2369.
- Hoffmann, M.; Krüger, N.; Schulz, S.; Cossmann, A.; Rocha, C.; Kempf, A.; Nehlmeier, I.; Graichen, L.; Moldenhauer, A.S.; Winkler, M.S.; et al. The Omicron variant is highly resistant against antibody-mediated neutralization: Implications for control of the COVID-19 pandemic. Cell 2022, 185, 447–456.e411.
- Kim, S.J.; Yao, Z.; Marsh, M.C.; Eckert, D.M.; Kay, M.S.; Lyakisheva, A.; Pasic, M.; Bansal, A.; Birnboim, C.; Jha, P.; et al. Homogeneous surrogate virus neutralization assay to rapidly assess neutralization activity of anti-SARS-CoV-2 antibodies. Nat. Commun. 2022, 13, 3716.
- Lauterio, A.; Valsecchi, M.; Santambrogio, S.; De Carlis, R.; Merli, M.; Calini, A.; Centonze, L.; Buscemi, V.; Bottiroli, M.; Puoti, M.; et al. Successful recovery from severe COVID-19 pneumonia after kidney transplantation: The interplay between immunosuppression and novel therapy including tocilizumab. Transpl. Infect. Dis. Off. J. Transplant. Soc. 2020, 22, e13334.
- Thompson, M.A.; Henderson, J.P.; Shah, P.K.; Rubinstein, S.M.; Joyner, M.J.; Choueiri, T.K.; Flora, D.B.; Griffiths, E.A.; Gulati, A.P.; Hwang, C.; et al. Association of Convalescent Plasma Therapy With Survival in Patients With Hematologic Cancers and COVID-19. JAMA Oncol. 2021, 7, 1167–1175.
- Betrains, A.; Godinas, L.; Woei, A.J.F.; Rosseels, W.; Van Herck, Y.; Lorent, N.; Dierickx, D.; Compernolle, V.; Meyfroidt, G.; Vanderbeke, L.; et al. Convalescent plasma treatment of persistent severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection in patients with lymphoma with impaired humoral immunity and lack of neutralising antibodies. Br. J. Haematol. 2021, 192, 1100–1105.
- Fung, M.; Nambiar, A.; Pandey, S.; Aldrich, J.M.; Teraoka, J.; Freise, C.; Roberts, J.; Chandran, S.; Hays, S.R.; Bainbridge, E.; et al. Treatment of immunocompromised COVID-19 patients with convalescent plasma. Transpl. Infect. Dis. Off. J. Transplant. Soc. 2021, 23, e13477.
- Ljungquist, O.; Lundgren, M.; Iliachenko, E.; Månsson, F.; Böttiger, B.; Landin-Olsson, M.; Wikén, C.; Rosendal, E.; Överby, A.K.; Wigren, B.J.; et al. Convalescent plasma treatment in severely immunosuppressed patients hospitalized with COVID-19: An observational study of 28 cases. Infect. Dis. 2022, 54, 283–291.
- Evolutionary insight into the emergence of SARS-CoV-2 variants of concern. Nat. Med. 2022, 28, 1357–1358.
- Clark, S.A.; Clark, L.E.; Pan, J.; Coscia, A.; McKay, L.G.A.; Shankar, S.; Johnson, R.I.; Brusic, V.; Choudhary, M.C.; Regan, J.; et al. SARS-CoV-2 evolution in an immunocompromised host reveals shared neutralization escape mechanisms. Cell 2021, 184, 2605–2617.e2618.
- Binnicker, M.J. Challenges and Controversies to Testing for COVID-19. J. Clin.Microbiol. 2020, 58, e01695-20.
- Zhao, J.; Liao, X.; Wang, H.; Wei, L.; Xing, M.; Liu, L.; Zhang, Z. Early virus clearance and delayed antibody response in a case of COVID-19 with a history of co-infection with HIV-1 and HCV. Clin. Infect. Dis. 2020, 71, 2233–2235.
- Xie, J.; Ding, C.; Li, J.; Wang, Y.; Guo, H.; Lu, Z.; Wang, J.; Zheng, C.; Jin, T.; Gao, Y. Characteristics of patients with coronavirus disease (COVID-19) confirmed using an IgM-IgG antibody test. J. Med. Virol. 2020, 92, 2004–2010.
- Ismail, A.A. Serological tests for COVID-19 antibodies: Limitations must be recognized. Ann. Clin. Biochem. 2020, 57, 274–276.
