The DNA damage response (DDR) system plays an important role in maintaining genome stability and preventing related diseases. The DDR network comprises many proteins and posttranslational modifications (PTMs) to proteins, which work in a coordinated manner to counteract various genotoxic stresses. Lysine crotonylation (Kcr) is a newly identified PTM occurring in both core histone and non-histone proteins in various organisms. This novel PTM is classified as a reversible acylation modification, which is regulated by a variety of acylases and deacylases and the intracellular crotonyl-CoA substrate concentration. Kcr links cellular metabolism with gene regulation and is involved in numerous cellular processes.
- lysine crotonylation
- DNA damage response (DDR)
- DNA double-strand break (DSB)
1. Introduction
2. Kcr Is an Evolutionary Conserved and Abundant PTM
3. Enzymes Responsible for Reversible Kcr Regulation
4. Regulation of Kcr by the Cellular Concentrations of Crotonyl-CoA
5. Recognition of Kcr by Chromatin-Associated Proteins
6. The Emerging Role of Kcr in DSB Repair
This entry is adapted from the peer-reviewed paper 10.3390/biom12101428
References
- Hoeijmakers, J.H. DNA damage, aging, and cancer. N. Engl. J. Med. 2009, 361, 1475–1485.
- Varon, R.; Vissinga, C.; Platzer, M.; Cerosaletti, K.M.; Chrzanowska, K.H.; Saar, K.; Beckmann, G.; Seemanova, E.; Cooper, P.R.; Nowak, N.J.; et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell 1998, 93, 467–476.
- Savitsky, K.; Bar-Shira, A.; Gilad, S.; Rotman, G.; Ziv, Y.; Vanagaite, L.; Tagle, D.A.; Smith, S.; Uziel, T.; Sfez, S.; et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 1995, 268, 1749–1753.
- Ciccia, A.; Elledge, S.J. The DNA damage response: Making it safe to play with knives. Mol. Cell 2010, 40, 179–204.
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078.
- Wang, W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat. Rev. Genet. 2007, 8, 735–748.
- Harper, J.W.; Elledge, S.J. The DNA damage response: Ten years after. Mol. Cell 2007, 28, 739–745.
- Huen, M.S.; Chen, J. The DNA damage response pathways: At the crossroad of protein modifications. Cell Res. 2008, 18, 8–16.
- Gong, F.; Miller, K.M. Histone methylation and the DNA damage response. Mutat. Res. Rev. Mutat. Res. 2019, 780, 37–47.
- Wang, Z.; Zhu, W.G.; Xu, X. Ubiquitin-like modifications in the DNA damage response. Mutat. Res. 2017, 803–805, 56–75.
- Brown, J.S.; Jackson, S.P. Ubiquitylation, neddylation and the DNA damage response. Open Biol. 2015, 5, 150018.
- Hou, W.H.; Chen, S.H.; Yu, X. Poly-ADP ribosylation in DNA damage response and cancer therapy. Mutat. Res. Rev. Mutat. Res. 2019, 780, 82–91.
- Kolobynina, K.G.; Rapp, A.; Cardoso, M.C. Chromatin Ubiquitination Guides DNA Double Strand Break Signaling and Repair. Front Cell Dev. Biol. 2022, 10, 928113.
- Kim, J.J.; Lee, S.Y.; Miller, K.M. Preserving genome integrity and function: The DNA damage response and histone modifications. Crit. Rev. Biochem. Mol. Biol. 2019, 54, 208–241.
- Tang, J.; Cho, N.W.; Cui, G.; Manion, E.M.; Shanbhag, N.M.; Botuyan, M.V.; Mer, G.; Greenberg, R.A. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 2013, 20, 317–325.
- Jacquet, K.; Fradet-Turcotte, A.; Avvakumov, N.; Lambert, J.P.; Roques, C.; Pandita, R.K.; Paquet, E.; Herst, P.; Gingras, A.C.; Pandita, T.K.; et al. The TIP60 Complex Regulates Bivalent Chromatin Recognition by 53BP1 through Direct H4K20me Binding and H2AK15 Acetylation. Mol. Cell 2016, 62, 409–421.
- Miller, K.M.; Tjeertes, J.V.; Coates, J.; Legube, G.; Polo, S.E.; Britton, S.; Jackson, S.P. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 2010, 17, 1144–1151.
- Elia, A.E.; Boardman, A.P.; Wang, D.C.; Huttlin, E.L.; Everley, R.A.; Dephoure, N.; Zhou, C.; Koren, I.; Gygi, S.P.; Elledge, S.J. Quantitative Proteomic Atlas of Ubiquitination and Acetylation in the DNA Damage Response. Mol. Cell 2015, 59, 867–881.
- Gong, F.; Chiu, L.Y.; Miller, K.M. Acetylation Reader Proteins: Linking Acetylation Signaling to Genome Maintenance and Cancer. PLoS Genet 2016, 12, e1006272.
- Sabari, B.R.; Zhang, D.; Allis, C.D.; Zhao, Y. Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol. 2017, 18, 90–101.
- Huang, H.; Sabari, B.R.; Garcia, B.A.; Allis, C.D.; Zhao, Y. SnapShot: Histone modifications. Cell 2014, 159, 458.
- Chen, Y.; Sprung, R.; Tang, Y.; Ball, H.; Sangras, B.; Kim, S.C.; Falck, J.R.; Peng, J.; Gu, W.; Zhao, Y. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell Proteomics 2007, 6, 812–819.
- Dai, L.; Peng, C.; Montellier, E.; Lu, Z.; Chen, Y.; Ishii, H.; Debernardi, A.; Buchou, T.; Rousseaux, S.; Jin, F.; et al. Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nat. Chem. Biol. 2014, 10, 365–370.
- Xie, Z.; Dai, J.; Dai, L.; Tan, M.; Cheng, Z.; Wu, Y.; Boeke, J.D.; Zhao, Y. Lysine succinylation and lysine malonylation in histones. Mol. Cell Proteomics 2012, 11, 100–107.
- Tan, M.; Peng, C.; Anderson, K.A.; Chhoy, P.; Xie, Z.; Dai, L.; Park, J.; Chen, Y.; Huang, H.; Zhang, Y.; et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 2014, 19, 605–617.
- Tan, M.; Luo, H.; Lee, S.; Jin, F.; Yang, J.S.; Montellier, E.; Buchou, T.; Cheng, Z.; Rousseaux, S.; Rajagopal, N.; et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 2011, 146, 1016–1028.
- Xie, Z.; Zhang, D.; Chung, D.; Tang, Z.; Huang, H.; Dai, L.; Qi, S.; Li, J.; Colak, G.; Chen, Y.; et al. Metabolic Regulation of Gene Expression by Histone Lysine beta-Hydroxybutyrylation. Mol. Cell 2016, 62, 194–206.
- Jiang, G.; Nguyen, D.; Archin, N.M.; Yukl, S.A.; Mendez-Lagares, G.; Tang, Y.; Elsheikh, M.M.; Thompson, G.R., 3rd; Hartigan-O’Connor, D.J.; Margolis, D.M.; et al. HIV latency is reversed by ACSS2-driven histone crotonylation. J. Clin. Investig. 2018, 128, 1190–1198.
- Ntorla, A.; Burgoyne, J.R. The Regulation and Function of Histone Crotonylation. Front. Cell Dev. Biol. 2021, 9, 624914.
- Wan, J.; Liu, H.; Chu, J.; Zhang, H. Functions and mechanisms of lysine crotonylation. J. Cell Mol. Med. 2019, 23, 7163–7169.
- Ruiz-Andres, O.; Sanchez-Nino, M.D.; Cannata-Ortiz, P.; Ruiz-Ortega, M.; Egido, J.; Ortiz, A.; Sanz, A.B. Histone lysine crotonylation during acute kidney injury in mice. Dis. Model Mech. 2016, 9, 633–645.
- Li, K.; Wang, Z. Histone crotonylation-centric gene regulation. Epigenetics Chromatin 2021, 14, 10.
- Hou, J.Y.; Zhou, L.; Li, J.L.; Wang, D.P.; Cao, J.M. Emerging roles of non-histone protein crotonylation in biomedicine. Cell Biosci. 2021, 11, 101.
- Xu, W.; Wan, J.; Zhan, J.; Li, X.; He, H.; Shi, Z.; Zhang, H. Global profiling of crotonylation on non-histone proteins. Cell Res. 2017, 27, 946–949.
- Kwon, O.K.; Kim, S.J.; Lee, S. First profiling of lysine crotonylation of myofilament proteins and ribosomal proteins in zebrafish embryos. Sci. Rep. 2018, 8, 3652.
- Chen, W.; Tang, D.; Xu, Y.; Zou, Y.; Sui, W.; Dai, Y.; Diao, H. Comprehensive analysis of lysine crotonylation in proteome of maintenance hemodialysis patients. Medicine 2018, 97, e12035.
- Sun, J.; Qiu, C.; Qian, W.; Wang, Y.; Sun, L.; Li, Y.; Ding, Z. Ammonium triggered the response mechanism of lysine crotonylome in tea plants. BMC Genomics 2019, 20, 340.
- Lin, H.; Tang, D.; Xu, Y.; Zhang, R.; Ou, M.; Zheng, F.; Chen, J.; Zhang, Y.; Zou, G.; Xue, W.; et al. Quantitative analysis of protein crotonylation identifies its association with immunoglobulin A nephropathy. Mol. Med. Rep. 2020, 21, 1242–1250.
- Liu, K.; Yuan, C.; Li, H.; Chen, K.; Lu, L.; Shen, C.; Zheng, X. A qualitative proteome-wide lysine crotonylation profiling of papaya (Carica papaya L.). Sci. Rep. 2018, 8, 8230.
- Liu, S.; Xue, C.; Fang, Y.; Chen, G.; Peng, X.; Zhou, Y.; Chen, C.; Liu, G.; Gu, M.; Wang, K.; et al. Global Involvement of Lysine Crotonylation in Protein Modification and Transcription Regulation in Rice. Mol. Cell Proteomics 2018, 17, 1922–1936.
- Yin, D.; Jiang, N.; Zhang, Y.; Wang, D.; Sang, X.; Feng, Y.; Chen, R.; Wang, X.; Yang, N.; Chen, Q. Global Lysine Crotonylation and 2-Hydroxyisobutyrylation in Phenotypically Different Toxoplasma gondii Parasites. Mol. Cell Proteomics 2019, 18, 2207–2224.
- Grunstein, M. Histone acetylation in chromatin structure and transcription. Nature 1997, 389, 349–352.
- Bonnaud, E.M.; Suberbielle, E.; Malnou, C.E. Histone acetylation in neuronal (dys)function. Biomol. Concepts 2016, 7, 103–116.
- Koprinarova, M.; Schnekenburger, M.; Diederich, M. Role of Histone Acetylation in Cell Cycle Regulation. Curr. Top Med. Chem. 2016, 16, 732–744.
- Choi, J.K.; Howe, L.J. Histone acetylation: Truth of consequences? Biochem. Cell Biol. 2009, 87, 139–150.
- Wade, P.A.; Pruss, D.; Wolffe, A.P. Histone acetylation: Chromatin in action. Trends Biochem. Sci. 1997, 22, 128–132.
- Gong, F.; Miller, K.M. Mammalian DNA repair: HATs and HDACs make their mark through histone acetylation. Mutat. Res. 2013, 750, 23–30.
- Roth, S.Y.; Denu, J.M.; Allis, C.D. Histone acetyltransferases. Annu. Rev. Biochem. 2001, 70, 81–120.
- Ogryzko, V.V. Mammalian histone acetyltransferases and their complexes. Cell Mol. Life Sci. 2001, 58, 683–692.
- Marmorstein, R.; Roth, S.Y. Histone acetyltransferases: Function, structure, and catalysis. Curr. Opin. Genet. Dev. 2001, 11, 155–161.
- Marmorstein, R. Structure and function of histone acetyltransferases. Cell Mol Life Sci 2001, 58, 693–703.
- Aka, J.A.; Kim, G.W.; Yang, X.J. K-acetylation and its enzymes: Overview and new developments. Handb. Exp. Pharmacol. 2011, 206, 1–12.
- Chen, H.P.; Zhao, Y.T.; Zhao, T.C. Histone deacetylases and mechanisms of regulation of gene expression. Crit. Rev. Oncog. 2015, 20, 35–47.
- De Ruijter, A.J.; van Gennip, A.H.; Caron, H.N.; Kemp, S.; van Kuilenburg, A.B. Histone deacetylases (HDACs): Characterization of the classical HDAC family. Biochem. J. 2003, 370, 737–749.
- Seto, E.; Yoshida, M. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6, a018713.
- Shvedunova, M.; Akhtar, A. Modulation of cellular processes by histone and non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 2022, 23, 329–349.
- Zhao, S.; Zhang, X.; Li, H. Beyond histone acetylation-writing and erasing histone acylations. Curr. Opin. Struct. Biol. 2018, 53, 169–177.
- Sabari, B.R.; Tang, Z.; Huang, H.; Yong-Gonzalez, V.; Molina, H.; Kong, H.E.; Dai, L.; Shimada, M.; Cross, J.R.; Zhao, Y.; et al. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol. Cell 2015, 58, 203–215.
- Liu, X.; Wei, W.; Liu, Y.; Yang, X.; Wu, J.; Zhang, Y.; Zhang, Q.; Shi, T.; Du, J.X.; Zhao, Y.; et al. MOF as an evolutionarily conserved histone crotonyltransferase and transcriptional activation by histone acetyltransferase-deficient and crotonyltransferase-competent CBP/p300. Cell Discov. 2017, 3, 17016.
- Kollenstart, L.; de Groot, A.J.L.; Janssen, G.M.C.; Cheng, X.; Vreeken, K.; Martino, F.; Cote, J.; van Veelen, P.A.; van Attikum, H. Gcn5 and Esa1 function as histone crotonyltransferases to regulate crotonylation-dependent transcription. J. Biol. Chem. 2019, 294, 20122–20134.
- Xiao, Y.; Li, W.; Yang, H.; Pan, L.; Zhang, L.; Lu, L.; Chen, J.; Wei, W.; Ye, J.; Li, J.; et al. HBO1 is a versatile histone acyltransferase critical for promoter histone acylations. Nucleic Acids Res. 2021, 49, 8037–8059.
- Yang, X.J.; Seto, E. The Rpd3/Hda1 family of lysine deacetylases: From bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 2008, 9, 206–218.
- Houtkooper, R.H.; Pirinen, E.; Auwerx, J. Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 2012, 13, 225–238.
- Wei, W.; Liu, X.; Chen, J.; Gao, S.; Lu, L.; Zhang, H.; Ding, G.; Wang, Z.; Chen, Z.; Shi, T.; et al. Class I histone deacetylases are major histone decrotonylases: Evidence for critical and broad function of histone crotonylation in transcription. Cell Res. 2017, 27, 898–915.
- Kelly, R.D.W.; Chandru, A.; Watson, P.J.; Song, Y.; Blades, M.; Robertson, N.S.; Jamieson, A.G.; Schwabe, J.W.R.; Cowley, S.M. Histone deacetylase (HDAC) 1 and 2 complexes regulate both histone acetylation and crotonylation in vivo. Sci. Rep. 2018, 8, 14690.
- Hao, S.; Wang, Y.; Zhao, Y.; Gao, W.; Cui, W.; Li, Y.; Cui, J.; Liu, Y.; Lin, L.; Xu, X.; et al. Dynamic switching of crotonylation to ubiquitination of H2A at lysine 119 attenuates transcription-replication conflicts caused by replication stress. Nucleic Acids Res. 2022, 50, 9873–9892.
- Bao, X.; Wang, Y.; Li, X.; Li, X.M.; Liu, Z.; Yang, T.; Wong, C.F.; Zhang, J.; Hao, Q.; Li, X.D. Identification of ’erasers’ for lysine crotonylated histone marks using a chemical proteomics approach. Elife 2014, 3, e02999.
- Fellows, R.; Denizot, J.; Stellato, C.; Cuomo, A.; Jain, P.; Stoyanova, E.; Balazsi, S.; Hajnady, Z.; Liebert, A.; Kazakevych, J.; et al. Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat. Commun. 2018, 9, 105.
- Wagner, G.R.; Hirschey, M.D. Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol. Cell 2014, 54, 5–16.
- Baddiley, J.; Kekwick, R.A.; Thain, E.M. A new method for acetylating proteins. Nature 1952, 170, 968–970.
- Liu, S.; Yu, H.; Liu, Y.; Liu, X.; Zhang, Y.; Bu, C.; Yuan, S.; Chen, Z.; Xie, G.; Li, W.; et al. Chromodomain Protein CDYL Acts as a Crotonyl-CoA Hydratase to Regulate Histone Crotonylation and Spermatogenesis. Mol. Cell 2017, 67, 853–866.e5.
- Fang, Y.; Xu, X.; Ding, J.; Yang, L.; Doan, M.T.; Karmaus, P.W.F.; Snyder, N.W.; Zhao, Y.; Li, J.L.; Li, X. Histone crotonylation promotes mesoendodermal commitment of human embryonic stem cells. Cell Stem Cell 2021, 28, 748–763.e7.
- Baumann, K. Post-translational modifications: Crotonylation versus acetylation. Nat. Rev. Mol. Cell Biol. 2015, 16, 265.
- Li, Y.; Sabari, B.R.; Panchenko, T.; Wen, H.; Zhao, D.; Guan, H.; Wan, L.; Huang, H.; Tang, Z.; Zhao, Y.; et al. Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain. Mol. Cell 2016, 62, 181–193.
- Andrews, F.H.; Shinsky, S.A.; Shanle, E.K.; Bridgers, J.B.; Gest, A.; Tsun, I.K.; Krajewski, K.; Shi, X.; Strahl, B.D.; Kutateladze, T.G. The Taf14 YEATS domain is a reader of histone crotonylation. Nat. Chem. Biol. 2016, 12, 396–398.
- Zhao, D.; Guan, H.; Zhao, S.; Mi, W.; Wen, H.; Li, Y.; Zhao, Y.; Allis, C.D.; Shi, X.; Li, H. YEATS2 is a selective histone crotonylation reader. Cell Res. 2016, 26, 629–632.
- Schulze, J.M.; Wang, A.Y.; Kobor, M.S. YEATS domain proteins: A diverse family with many links to chromatin modification and transcription. Biochem. Cell Biol. 2009, 87, 65–75.
- Li, Y.; Wen, H.; Xi, Y.; Tanaka, K.; Wang, H.; Peng, D.; Ren, Y.; Jin, Q.; Dent, S.Y.; Li, W.; et al. AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation. Cell 2014, 159, 558–571.
- Xiong, X.; Panchenko, T.; Yang, S.; Zhao, S.; Yan, P.; Zhang, W.; Xie, W.; Li, Y.; Zhao, Y.; Allis, C.D.; et al. Selective recognition of histone crotonylation by double PHD fingers of MOZ and DPF2. Nat. Chem. Biol. 2016, 12, 1111–1118.
- Klein, B.J.; Jang, S.M.; Lachance, C.; Mi, W.; Lyu, J.; Sakuraba, S.; Krajewski, K.; Wang, W.W.; Sidoli, S.; Liu, J.; et al. Histone H3K23-specific acetylation by MORF is coupled to H3K14 acylation. Nat. Commun. 2019, 10, 4724.
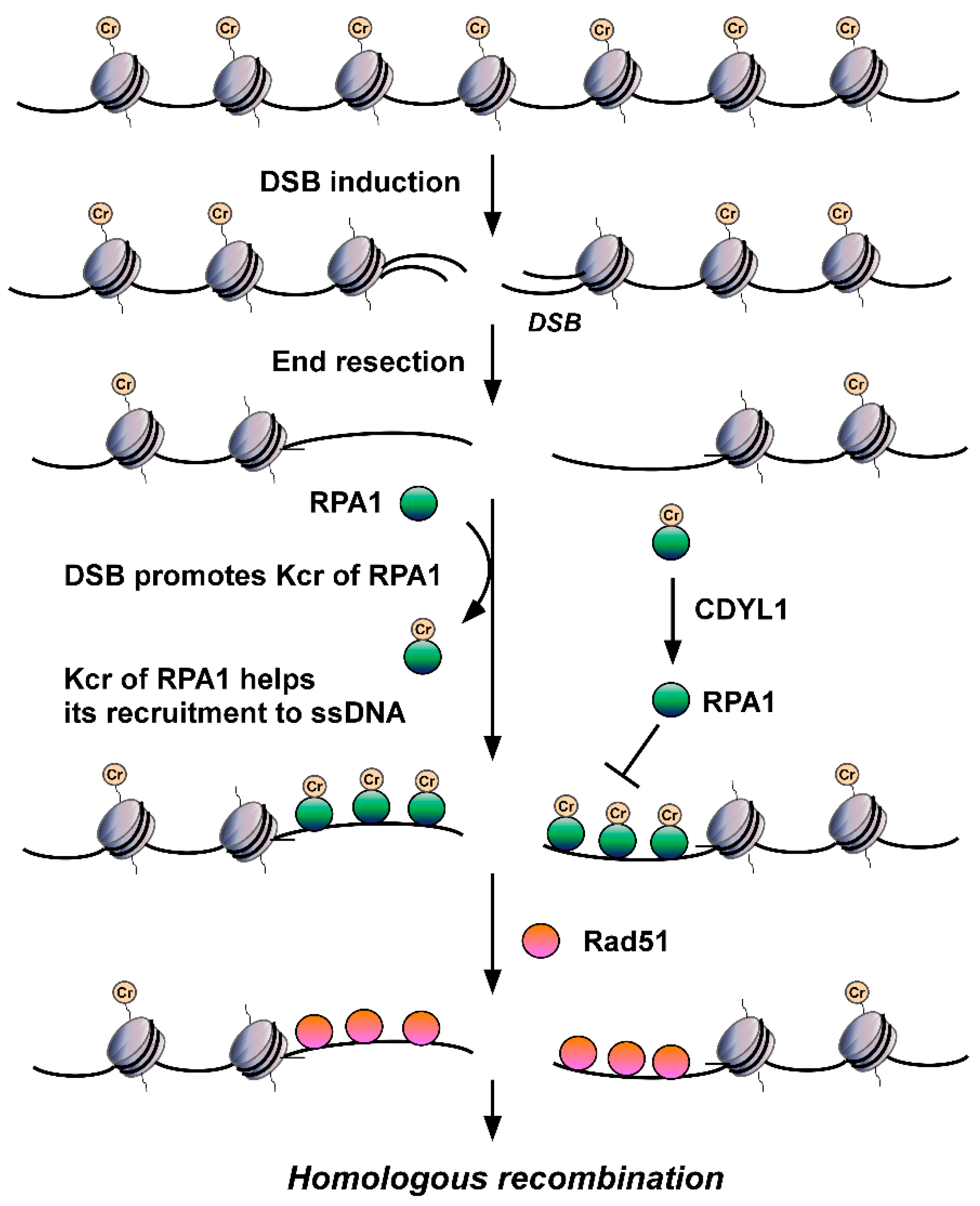
- Yu, H.; Bu, C.; Liu, Y.; Gong, T.; Liu, X.; Liu, S.; Peng, X.; Zhang, W.; Peng, Y.; Yang, J.; et al. Global crotonylome reveals CDYL-regulated RPA1 crotonylation in homologous recombination-mediated DNA repair. Sci. Adv. 2020, 6, eaay4697.
- Abu-Zhayia, E.R.; Bishara, L.A.; Machour, F.E.; Barisaac, A.S.; Ben-Oz, B.M.; Ayoub, N. CDYL1-dependent decrease in lysine crotonylation at DNA double-strand break sites functionally uncouples transcriptional silencing and repair. Mol. Cell 2022, 82, 1940–1955.e7.
