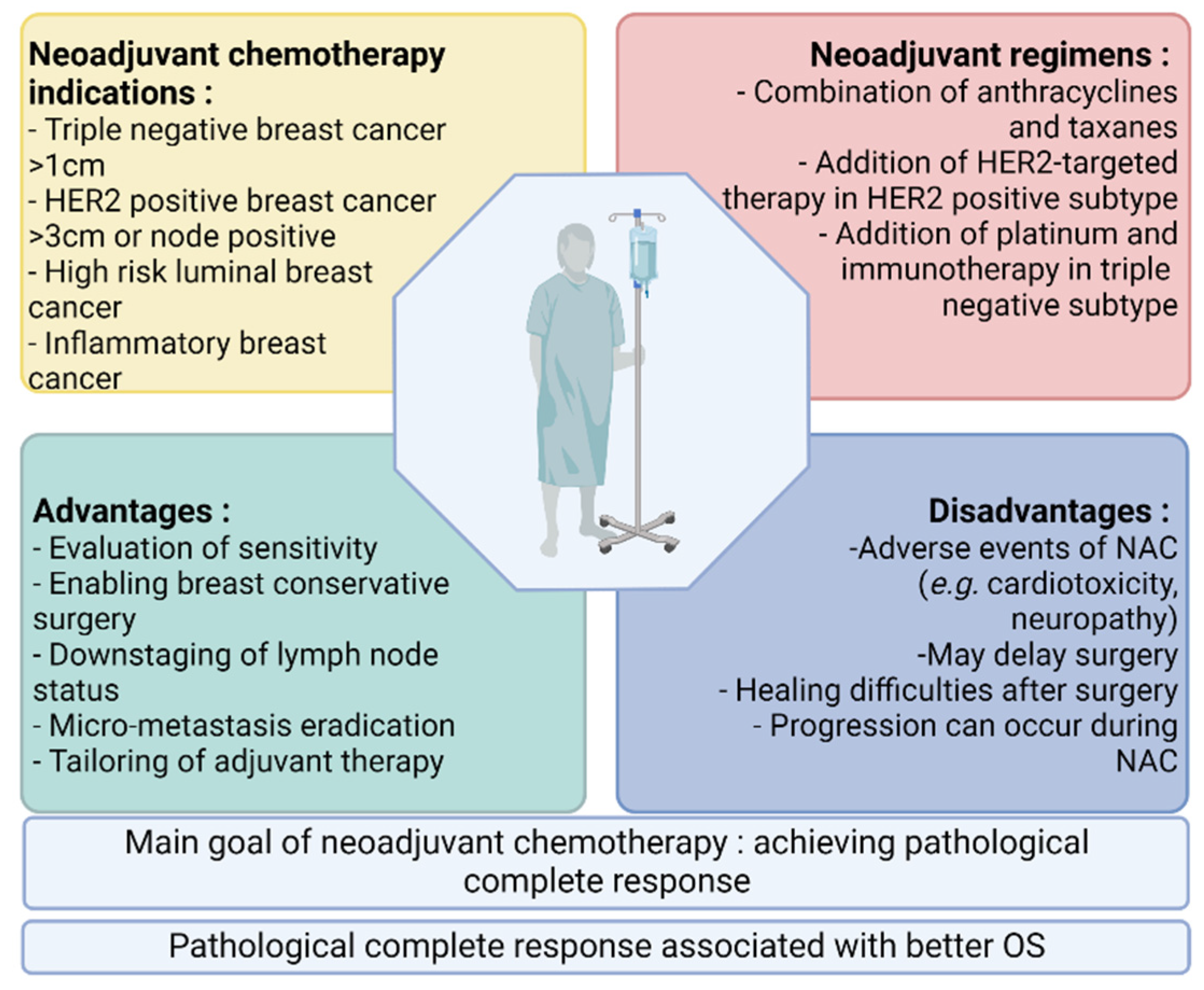
Pathological complete response (pCR) after neoadjuvant chemotherapy in patients with early breast cancer is correlated with better survival. Meanwhile, an expanding arsenal of post-neoadjuvant treatment strategies have proven beneficial in the absence of pCR, leading to an increased use of neoadjuvant systemic therapy in patients with early breast cancer and the search for predictive biomarkers of response. The better prediction of response to neoadjuvant chemotherapy could enable the escalation or de-escalation of neoadjuvant treatment strategies, with the ultimate goal of improving the clinical management of early breast cancer.
- breast cancer
- neoadjuvant chemotherapy
- biomarkers
- predictive factors
1. Introduction
2. Breast Cancer Subtypes and Intratumoral Heterogeneity
2.1. Molecular Classification and Intrinsic Subtypes
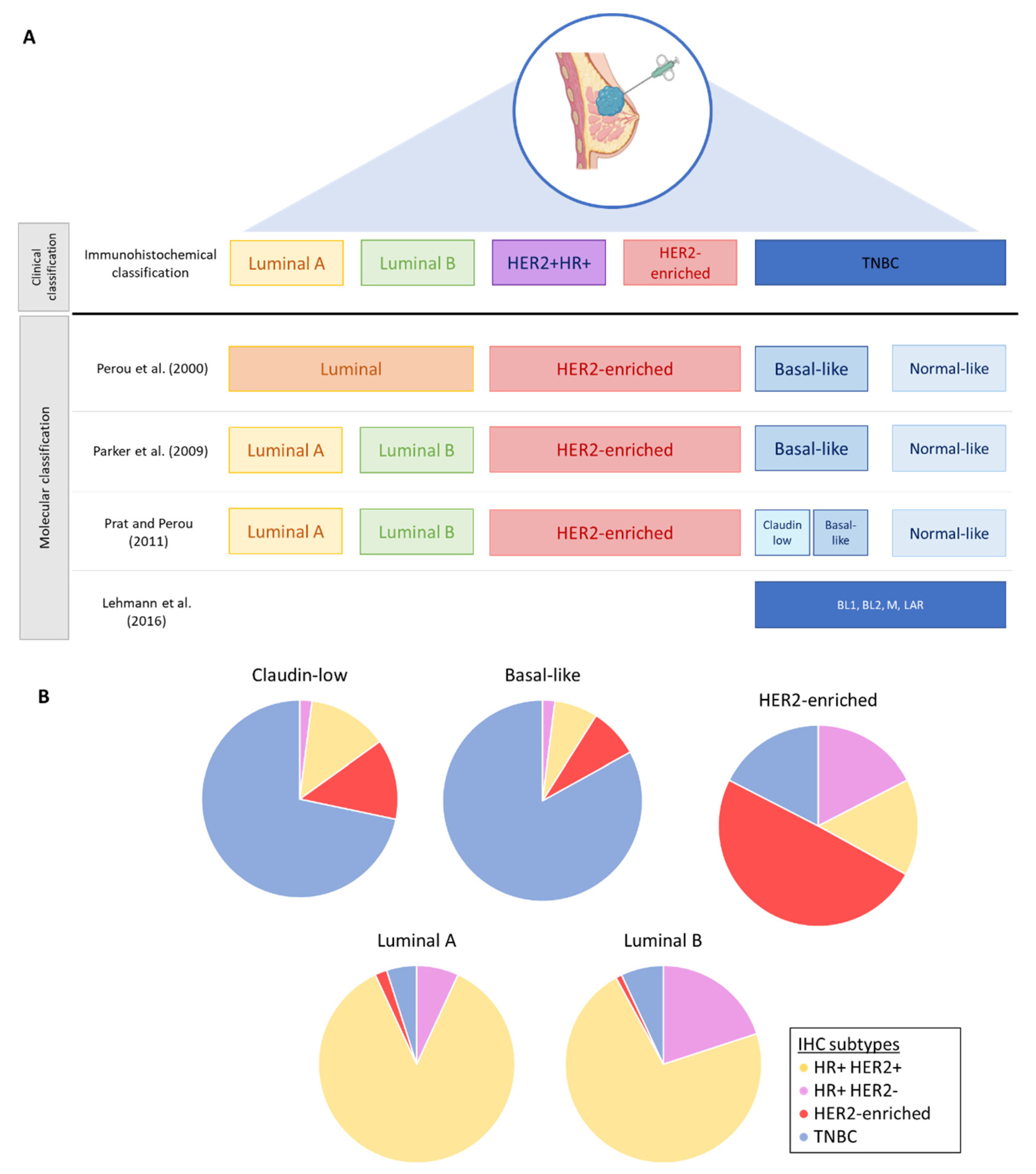
2.2. Intratumoral Heterogeneity in Breast Cancers
3. Resistance to Neoadjuvant Chemotherapy
3.1. Drug-Associated Resistance
3.2. Cancer Cell-Associated Resistance
4. Current Biomarkers Used for the Clinical Decision Making of Breast Cancer Patients
4.1. Ki-67 before NAC
4.2. Tumor Size
4.3. Surrogate Molecular Subtypes as Determined by Immunohistochemistry
4.4. Tumor-Infiltrating Lymphocytes (TILs)
4.5. PD-L1 Expression
5. Predictive Biomarkers under Investigation
5.1. Imaging and Radiomics Biomarkers
5.1.1. MRI
5.1.2. Quantitative Ultrasound
Compared to MRI, ultrasound imaging has several advantages such as its lower cost, the absence of the injection of exogenous contrast agents and the fact that it is transportable. It is therefore more accessible for the screening and evaluation of all patients. QUS is a technique that extracts characteristics of the physical properties of tissues (e.g., elastography) both in intratumoral and marginal regions. Different studies have evaluated the evolution in the structure of the tumor tissue after treatment by QUS. This technique can detect tumor cell death in response to chemotherapy and, in addition, could predict response to NAC after one-to-four weeks of chemotherapy [60][61][62][63][64][65].
5.1.3. 18F-FDG PET/CT
5.2. Plasmatic Biomarkers
5.2.1. Peripheral Blood Cells and Ratios
5.2.2. Liquid Biopsies
5.3. Gene Signatures
5.3.1. EndoPredict—Molecular Score (MS)
5.3.2. Oncotype DX—Recurrence Score (RS)
5.3.3. Mammaprint
5.3.4. PAM50—Prosigna Assay
6. Conclusions
This entry is adapted from the peer-reviewed paper 10.3390/cancers14163876
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249.
- Loi, S. The ESMO clinical practise guidelines for early breast cancer: Diagnosis, treatment and follow-up: On the winding road to personalized medicine. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2019, 30, 1183–1184.
- Cardoso, F.; Kyriakides, S.; Ohno, S.; Penault-Llorca, F.; Poortmans, P.; Rubio, I.T.; Zackrisson, S.; Senkus, E. Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2019, 30, 1194–1220.
- Cardoso, F.; Paluch-Shimon, S.; Senkus, E.; Curigliano, G.; Aapro, M.S.; André, F.; Barrios, C.H.; Bergh, J.; Bhattacharyya, G.S.; Biganzoli, L.; et al. 5th ESO-ESMO international consensus guidelines for advanced breast cancer (ABC 5). Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2020, 31, 1623–1649.
- Mauri, D.; Pavlidis, N.; Ioannidis, J.P. Neoadjuvant versus adjuvant systemic treatment in breast cancer: A meta-analysis. J. Natl. Cancer Inst. 2005, 97, 188–194.
- Charfare, H.; Limongelli, S.; Purushotham, A.D. Neoadjuvant chemotherapy in breast cancer. Br. J. Surg. 2005, 92, 14–23.
- von Minckwitz, G.; Untch, M.; Blohmer, J.U.; Costa, S.D.; Eidtmann, H.; Fasching, P.A.; Gerber, B.; Eiermann, W.; Hilfrich, J.; Huober, J.; et al. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J. Clin. Oncol. 2012, 30, 1796–1804.
- Asaoka, M.; Gandhi, S.; Ishikawa, T.; Takabe, K. Neoadjuvant Chemotherapy for Breast Cancer: Past, Present, and Future. Breast Cancer Basic Clin. Res. 2020, 14, 1178223420980377.
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752.
- Parker, J.S.; Mullins, M.; Cheang, M.C.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009, 27, 1160–1167.
- Sørlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874.
- Hugh, J.; Hanson, J.; Cheang, M.C.; Nielsen, T.O.; Perou, C.M.; Dumontet, C.; Reed, J.; Krajewska, M.; Treilleux, I.; Rupin, M.; et al. Breast cancer subtypes and response to docetaxel in node-positive breast cancer: Use of an immunohistochemical definition in the BCIRG 001 trial. J. Clin. Oncol. 2009, 27, 1168–1176.
- Carey, L.A.; Berry, D.A.; Cirrincione, C.T.; Barry, W.T.; Pitcher, B.N.; Harris, L.N.; Ollila, D.W.; Krop, I.E.; Henry, N.L.; Weckstein, D.J.; et al. Molecular Heterogeneity and Response to Neoadjuvant Human Epidermal Growth Factor Receptor 2 Targeting in CALGB 40601, a Randomized Phase III Trial of Paclitaxel Plus Trastuzumab With or Without Lapatinib. J. Clin. Oncol. 2016, 34, 542–549.
- Rouzier, R.; Perou, C.M.; Symmans, W.F.; Ibrahim, N.; Cristofanilli, M.; Anderson, K.; Hess, K.R.; Stec, J.; Ayers, M.; Wagner, P.; et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2005, 11, 5678–5685.
- Marra, A.; Trapani, D.; Viale, G.; Criscitiello, C.; Curigliano, G. Practical classification of triple-negative breast cancer: Intratumoral heterogeneity, mechanisms of drug resistance, and novel therapies. NPJ Breast Cancer 2020, 6, 54.
- Prat, A.; Perou, C.M. Deconstructing the molecular portraits of breast cancer. Mol. Oncol. 2011, 5, 5–23.
- Lehmann, B.D.; Jovanović, B.; Chen, X.; Estrada, M.V.; Johnson, K.N.; Shyr, Y.; Moses, H.L.; Sanders, M.E.; Pietenpol, J.A. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS ONE 2016, 11, e0157368.
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767.
- Haynes, B.; Sarma, A.; Nangia-Makker, P.; Shekhar, M.P. Breast cancer complexity: Implications of intratumoral heterogeneity in clinical management. Cancer Metastasis Rev. 2017, 36, 547–555.
- Zardavas, D.; Irrthum, A.; Swanton, C.; Piccart, M. Clinical management of breast cancer heterogeneity. Nat. Rev. Clin. Oncol. 2015, 12, 381–394.
- Lüönd, F.; Tiede, S.; Christofori, G. Breast cancer as an example of tumour heterogeneity and tumour cell plasticity during malignant progression. Br. J. Cancer 2021, 125, 164–175.
- Turner, K.M.; Yeo, S.K.; Holm, T.M.; Shaughnessy, E.; Guan, J.L. Heterogeneity within molecular subtypes of breast cancer. Am. J. Physiol. Cell Physiol. 2021, 321, C343–C354.
- Tuasha, N.; Petros, B. Heterogeneity of Tumors in Breast Cancer: Implications and Prospects for Prognosis and Therapeutics. Scientifica 2020, 2020, 4736091.
- Zhou, S.; Huang, Y.-E.; Liu, H.; Zhou, X.; Yuan, M.; Hou, F.; Wang, L.; Jiang, W. Single-cell RNA-seq dissects the intratumoral heterogeneity of triple-negative breast cancer based on gene regulatory networks. Mol. Ther.-Nucleic Acids 2021, 23, 682–690.
- Luo, B.; Yan, D.; Yan, H.; Yuan, J. Cytochrome P450: Implications for human breast cancer (Review). Oncol. Lett. 2021, 22, 548.
- Bray, J.; Sludden, J.; Griffin, M.J.; Cole, M.; Verrill, M.; Jamieson, D.; Boddy, A.V. Influence of pharmacogenetics on response and toxicity in breast cancer patients treated with doxorubicin and cyclophosphamide. Br. J. Cancer 2010, 102, 1003–1009.
- Marsh, S.; Somlo, G.; Li, X.; Frankel, P.; King, C.R.; Shannon, W.D.; McLeod, H.L.; Synold, T.W. Pharmacogenetic analysis of paclitaxel transport and metabolism genes in breast cancer. Pharm. J. 2007, 7, 362–365.
- Seredina, T.A.; Goreva, O.B.; Talaban, V.O.; Grishanova, A.Y.; Lyakhovich, V.V. Association of cytochrome P450 genetic polymorphisms with neoadjuvant chemotherapy efficacy in breast cancer patients. BMC Med. Genet. 2012, 13, 45.
- An, J.; Peng, C.; Tang, H.; Liu, X.; Peng, F. New Advances in the Research of Resistance to Neoadjuvant Chemotherapy in Breast Cancer. Int. J. Mol. Sci. 2021, 22, 9644.
- Ferrari, P.; Scatena, C.; Ghilli, M.; Bargagna, I.; Lorenzini, G.; Nicolini, A. Molecular Mechanisms, Biomarkers and Emerging Therapies for Chemotherapy Resistant TNBC. Int. J. Mol. Sci. 2022, 23, 1665.
- Lee, H.E.; Kim, J.H.; Kim, Y.J.; Choi, S.Y.; Kim, S.W.; Kang, E.; Chung, I.Y.; Kim, I.A.; Kim, E.J.; Choi, Y.; et al. An increase in cancer stem cell population after primary systemic therapy is a poor prognostic factor in breast cancer. Br. J. Cancer 2011, 104, 1730–1738.
- Park, S.Y.; Lee, H.E.; Li, H.; Shipitsin, M.; Gelman, R.; Polyak, K. Heterogeneity for stem cell-related markers according to tumor subtype and histologic stage in breast cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2010, 16, 876–887.
- Ma, F.; Li, H.; Wang, H.; Shi, X.; Fan, Y.; Ding, X.; Lin, C.; Zhan, Q.; Qian, H.; Xu, B. Enriched CD44+/CD24− population drives the aggressive phenotypes presented in triple-negative breast cancer (TNBC). Cancer Lett. 2014, 353, 153–159.
- Wang, H.; Wang, L.; Song, Y.; Wang, S.; Huang, X.; Xuan, Q.; Kang, X.; Zhang, Q. CD44(+)/CD24(-) phenotype predicts a poor prognosis in triple-negative breast cancer. Oncol. Lett. 2017, 14, 5890–5898.
- Zong, B.; Sun, L.; Peng, Y.; Wang, Y.; Yu, Y.; Lei, J.; Zhang, Y.; Guo, S.; Li, K.; Liu, S. HORMAD1 promotes docetaxel resistance in triple negative breast cancer by enhancing DNA damage tolerance Corrigendum in /10.3892/or.2021.8146. Oncol. Rep. 2021, 46, 138.
- Smith, B.N.; Bhowmick, N.A. Role of EMT in Metastasis and Therapy Resistance. J. Clin. Med. 2016, 5, 17.
- Wang, Y.; Wang, X.; Zhao, H.; Liang, B.; Du, Q. Clusterin confers resistance to TNF-alpha-induced apoptosis in breast cancer cells through NF-kappaB activation and Bcl-2 overexpression. J. Chemother. 2012, 24, 348–357.
- Ozretic, P.; Alvir, I.; Sarcevic, B.; Vujaskovic, Z.; Rendic-Miocevic, Z.; Roguljic, A.; Beketic-Oreskovic, L. Apoptosis regulator Bcl-2 is an independent prognostic marker for worse overall survival in triple-negative breast cancer patients. Int. J. Biol. Markers 2018, 33, 109–115.
- Campbell, K.J.; Dhayade, S.; Ferrari, N.; Sims, A.H.; Johnson, E.; Mason, S.M.; Dickson, A.; Ryan, K.M.; Kalna, G.; Edwards, J.; et al. MCL-1 is a prognostic indicator and drug target in breast cancer. Cell Death Dis. 2018, 9, 19.
- Li, L.; Han, D.; Wang, X.; Wang, Q.; Tian, J.; Yao, J.; Yuan, L.; Qian, K.; Zou, Q.; Yi, W.; et al. Prognostic values of Ki-67 in neoadjuvant setting for breast cancer: A systematic review and meta-analysis. Future Oncol. 2017, 13, 1021–1034.
- Scholzen, T.; Gerdes, J. The Ki-67 protein: From the known and the unknown. J. Cell. Physiol. 2000, 182, 311–322.
- Nielsen, T.O.; Leung, S.C.Y.; Rimm, D.L.; Dodson, A.; Acs, B.; Badve, S.; Denkert, C.; Ellis, M.J.; Fineberg, S.; Flowers, M.; et al. Assessment of Ki67 in Breast Cancer: Updated Recommendations From the International Ki67 in Breast Cancer Working Group. J. Natl. Cancer Inst. 2021, 113, 808–819.
- Chen, X.; He, C.; Han, D.; Zhou, M.; Wang, Q.; Tian, J.; Li, L.; Xu, F.; Zhou, E.; Yang, K. The predictive value of Ki-67 before neoadjuvant chemotherapy for breast cancer: A systematic review and meta-analysis. Future Oncol. 2017, 13, 843–857.
- Livingston-Rosanoff, D.; Schumacher, J.; Vande Walle, K.; Stankowski-Drengler, T.; Greenberg, C.C.; Neuman, H.; Wilke, L.G. Does Tumor Size Predict Response to Neoadjuvant Chemotherapy in the Modern Era of Biologically Driven Treatment? A Nationwide Study of US Breast Cancer Patients. Clin. Breast Cancer 2019, 19, e741–e747.
- Cortazar, P.; Zhang, L.; Untch, M.; Mehta, K.; Costantino, J.P.; Wolmark, N.; Bonnefoi, H.; Cameron, D.; Gianni, L.; Valagussa, P.; et al. Pathological complete response and long-term clinical benefit in breast cancer: The CTNeoBC pooled analysis. Lancet 2014, 384, 164–172.
- Haque, W.; Verma, V.; Hatch, S.; Suzanne Klimberg, V.; Brian Butler, E.; Teh, B.S. Response rates and pathologic complete response by breast cancer molecular subtype following neoadjuvant chemotherapy. Breast Cancer Res. Treat. 2018, 170, 559–567.
- Cortazar, P.; Geyer, C.E., Jr. Pathological complete response in neoadjuvant treatment of breast cancer. Ann. Surg. Oncol. 2015, 22, 1441–1446.
- Shen, G.; Zhao, F.; Huo, X.; Ren, D.; Du, F.; Zheng, F.; Zhao, J. Meta-Analysis of HER2-Enriched Subtype Predicting the Pathological Complete Response within HER2-Positive Breast Cancer in Patients Who Received Neoadjuvant Treatment. Front. Oncol. 2021, 11, 632357.
- von Minckwitz, G.; Untch, M.; Nüesch, E.; Loibl, S.; Kaufmann, M.; Kümmel, S.; Fasching, P.A.; Eiermann, W.; Blohmer, J.U.; Costa, S.D.; et al. Impact of treatment characteristics on response of different breast cancer phenotypes: Pooled analysis of the German neo-adjuvant chemotherapy trials. Breast Cancer Res. Treat. 2011, 125, 145–156.
- Solinas, C.; Ceppi, M.; Lambertini, M.; Scartozzi, M.; Buisseret, L.; Garaud, S.; Fumagalli, D.; de Azambuja, E.; Salgado, R.; Sotiriou, C.; et al. Tumor-infiltrating lymphocytes in patients with HER2-positive breast cancer treated with neoadjuvant chemotherapy plus trastuzumab, lapatinib or their combination: A meta-analysis of randomized controlled trials. Cancer Treat. Rev. 2017, 57, 8–15.
- Salgado, R.; Denkert, C.; Demaria, S.; Sirtaine, N.; Klauschen, F.; Pruneri, G.; Wienert, S.; Van den Eynden, G.; Baehner, F.L.; Penault-Llorca, F.; et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: Recommendations by an International TILs Working Group 2014. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2015, 26, 259–271.
- Hendry, S.; Salgado, R.; Gevaert, T.; Russell, P.A.; John, T.; Thapa, B.; Christie, M.; van de Vijver, K.; Estrada, M.V.; Gonzalez-Ericsson, P.I.; et al. Assessing Tumor-infiltrating Lymphocytes in Solid Tumors: A Practical Review for Pathologists and Proposal for a Standardized Method from the International Immunooncology Biomarkers Working Group: Part 1: Assessing the Host Immune Response, TILs in Invasive Breast Carcinoma and Ductal Carcinoma In Situ, Metastatic Tumor Deposits and Areas for Further Research. Adv. Anat. Pathol. 2017, 24, 235–251.
- Denkert, C.; Liedtke, C.; Tutt, A.; von Minckwitz, G. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet 2017, 389, 2430–2442.
- Vranic, S.; Cyprian, F.S.; Gatalica, Z.; Palazzo, J. PD-L1 status in breast cancer: Current view and perspectives. Semin. Cancer Biol. 2021, 72, 146–154.
- Marinelli, D.; Mazzotta, M.; Pizzuti, L.; Krasniqi, E.; Gamucci, T.; Natoli, C.; Grassadonia, A.; Tinari, N.; Tomao, S.; Sperduti, I.; et al. Neoadjuvant Immune-Checkpoint Blockade in Triple-Negative Breast Cancer: Current Evidence and Literature-Based Meta-Analysis of Randomized Trials. Cancers 2020, 12, 2497.
- Zhang, L.; Wang, X.I.; Ding, J.; Sun, Q.; Zhang, S. The predictive and prognostic value of Foxp3+/CD25+ regulatory T cells and PD-L1 expression in triple negative breast cancer. Ann. Diagn. Pathol. 2019, 40, 143–151.
- Chamming’s, F.; Ueno, Y.; Ferré, R.; Kao, E.; Jannot, A.S.; Chong, J.; Omeroglu, A.; Mesurolle, B.; Reinhold, C.; Gallix, B. Features from Computerized Texture Analysis of Breast Cancers at Pretreatment MR Imaging Are Associated with Response to Neoadjuvant Chemotherapy. Radiology 2018, 286, 412–420.
- Braman, N.; Prasanna, P.; Whitney, J.; Singh, S.; Beig, N.; Etesami, M.; Bates, D.D.B.; Gallagher, K.; Bloch, B.N.; Vulchi, M.; et al. Association of Peritumoral Radiomics with Tumor Biology and Pathologic Response to Preoperative Targeted Therapy for HER2 (ERBB2)-Positive Breast Cancer. JAMA Netw. Open 2019, 2, e192561.
- Liu, Z.; Li, Z.; Qu, J.; Zhang, R.; Zhou, X.; Li, L.; Sun, K.; Tang, Z.; Jiang, H.; Li, H.; et al. Radiomics of Multiparametric MRI for Pretreatment Prediction of Pathologic Complete Response to Neoadjuvant Chemotherapy in Breast Cancer: A Multicenter Study. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 3538–3547.
- Quiaoit, K.; DiCenzo, D.; Fatima, K.; Bhardwaj, D.; Sannachi, L.; Gangeh, M.; Sadeghi-Naini, A.; Dasgupta, A.; Kolios, M.C.; Trudeau, M.; et al. Quantitative ultrasound radiomics for therapy response monitoring in patients with locally advanced breast cancer: Multi-institutional study results. PLoS ONE 2020, 15, e0236182.
- Sadeghi-Naini, A.; Sannachi, L.; Pritchard, K.; Trudeau, M.; Gandhi, S.; Wright, F.C.; Zubovits, J.; Yaffe, M.J.; Kolios, M.C.; Czarnota, G.J. Early prediction of therapy responses and outcomes in breast cancer patients using quantitative ultrasound spectral texture. Oncotarget 2014, 5, 3497–3511.
- Tadayyon, H.; Sannachi, L.; Gangeh, M.J.; Kim, C.; Ghandi, S.; Trudeau, M.; Pritchard, K.; Tran, W.T.; Slodkowska, E.; Sadeghi-Naini, A.; et al. A priori Prediction of Neoadjuvant Chemotherapy Response and Survival in Breast Cancer Patients using Quantitative Ultrasound. Sci. Rep. 2017, 7, 45733.
- Sannachi, L.; Gangeh, M.; Tadayyon, H.; Gandhi, S.; Wright, F.C.; Slodkowska, E.; Curpen, B.; Sadeghi-Naini, A.; Tran, W.; Czarnota, G.J. Breast Cancer Treatment Response Monitoring Using Quantitative Ultrasound and Texture Analysis: Comparative Analysis of Analytical Models. Transl. Oncol. 2019, 12, 1271–1281.
- Fernandes, J.; Sannachi, L.; Tran, W.T.; Koven, A.; Watkins, E.; Hadizad, F.; Gandhi, S.; Wright, F.; Curpen, B.; El Kaffas, A.; et al. Monitoring Breast Cancer Response to Neoadjuvant Chemotherapy Using Ultrasound Strain Elastography. Transl. Oncol. 2019, 12, 1177–1184.
- Tadayyon, H.; Sannachi, L.; Gangeh, M.; Sadeghi-Naini, A.; Tran, W.; Trudeau, M.E.; Pritchard, K.; Ghandi, S.; Verma, S.; Czarnota, G.J. Quantitative ultrasound assessment of breast tumor response to chemotherapy using a multi-parameter approach. Oncotarget 2016, 7, 45094–45111.
- Paydary, K.; Seraj, S.M.; Zadeh, M.Z.; Emamzadehfard, S.; Shamchi, S.P.; Gholami, S.; Werner, T.J.; Alavi, A. The Evolving Role of FDG-PET/CT in the Diagnosis, Staging, and Treatment of Breast Cancer. Mol. Imaging Biol. 2019, 21, 1–10.
- Fowler, A.M.; Mankoff, D.A.; Joe, B.N. Imaging Neoadjuvant Therapy Response in Breast Cancer. Radiology 2017, 285, 358–375.
- Schelling, M.; Avril, N.; Nährig, J.; Kuhn, W.; Römer, W.; Sattler, D.; Werner, M.; Dose, J.; Jänicke, F.; Graeff, H.; et al. Positron emission tomography using Fluorodeoxyglucose for monitoring primary chemotherapy in breast cancer. J. Clin. Oncol. 2000, 18, 1689–1695.
- Lee, H.W.; Lee, H.M.; Choi, S.E.; Yoo, H.; Ahn, S.G.; Lee, M.K.; Jeong, J.; Jung, W.H. The Prognostic Impact of Early Change in 18F-FDG PET SUV After Neoadjuvant Chemotherapy in Patients with Locally Advanced Breast Cancer. J. Nucl. Med. Off. Publ. Soc. Nucl. Med. 2016, 57, 1183–1188.
- Dose Schwarz, J.; Bader, M.; Jenicke, L.; Hemminger, G.; Jänicke, F.; Avril, N. Early prediction of response to chemotherapy in metastatic breast cancer using sequential 18F-FDG PET. J. Nucl. Med. Off. Publ. Soc. Nucl. Med. 2005, 46, 1144–1150.
- Corbeau, I.; Jacot, W.; Guiu, S. Neutrophil to Lymphocyte Ratio as Prognostic and Predictive Factor in Breast Cancer Patients: A Systematic Review. Cancers 2020, 12, 958.
- Li, X.; Dai, D.; Chen, B.; Tang, H.; Xie, X.; Wei, W. The value of neutrophil-to-lymphocyte ratio for response and prognostic effect of neoadjuvant chemotherapy in solid tumors: A systematic review and meta-analysis. J. Cancer 2018, 9, 861–871.
- Guo, W.; Lu, X.; Liu, Q.; Zhang, T.; Li, P.; Qiao, W.; Deng, M. Prognostic value of neutrophil-to-lymphocyte ratio and platelet-to-lymphocyte ratio for breast cancer patients: An updated meta-analysis of 17079 individuals. Cancer Med. 2019, 8, 4135–4148.
- Chae, S.; Kang, K.M.; Kim, H.J.; Kang, E.; Park, S.Y.; Kim, J.H.; Kim, S.H.; Kim, S.W.; Kim, E.K. Neutrophil-lymphocyte ratio predicts response to chemotherapy in triple-negative breast cancer. Curr. Oncol. 2018, 25, e113–e119.
- Xue, L.B.; Liu, Y.H.; Zhang, B.; Yang, Y.F.; Yang, D.; Zhang, L.W.; Jin, J.; Li, J. Prognostic role of high neutrophil-to-lymphocyte ratio in breast cancer patients receiving neoadjuvant chemotherapy: Meta-analysis. Medicine 2019, 98, e13842.
- Cullinane, C.; Creavin, B.; O’Leary, D.P.; O’Sullivan, M.J.; Kelly, L.; Redmond, H.P.; Corrigan, M.A. Can the Neutrophil to Lymphocyte Ratio Predict Complete Pathologic Response to Neoadjuvant Breast Cancer Treatment? A Systematic Review and Meta-analysis. Clin. Breast Cancer 2020, 20, e675–e681.
- Zhu, J.; Jiao, D.; Zhao, Y.; Guo, X.; Yang, Y.; Xiao, H.; Liu, Z. Development of a predictive model utilizing the neutrophil to lymphocyte ratio to predict neoadjuvant chemotherapy efficacy in early breast cancer patients. Sci. Rep. 2021, 11, 1350.
- Magbanua, M.J.M.; Swigart, L.B.; Wu, H.T.; Hirst, G.L.; Yau, C.; Wolf, D.M.; Tin, A.; Salari, R.; Shchegrova, S.; Pawar, H.; et al. Circulating tumor DNA in neoadjuvant-treated breast cancer reflects response and survival. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2021, 32, 229–239.
- Beaver, J.A.; Jelovac, D.; Balukrishna, S.; Cochran, R.; Croessmann, S.; Zabransky, D.J.; Wong, H.Y.; Toro, P.V.; Cidado, J.; Blair, B.G.; et al. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014, 20, 2643–2650.
- Alimirzaie, S.; Bagherzadeh, M.; Akbari, M.R. Liquid biopsy in breast cancer: A comprehensive review. Clin. Genet. 2019, 95, 643–660.
- Shoukry, M.; Broccard, S.; Kaplan, J.; Gabriel, E. The Emerging Role of Circulating Tumor DNA in the Management of Breast Cancer. Cancers 2021, 13, 3813.
- Honoré, N.; Galot, R.; van Marcke, C.; Limaye, N.; Machiels, J.-P. Liquid Biopsy to Detect Minimal Residual Disease: Methodology and Impact. Cancers 2021, 13, 5364.
- Sant, M.; Bernat-Peguera, A.; Felip, E.; Margelí, M. Role of ctDNA in Breast Cancer. Cancers 2022, 14, 310.
- Filipits, M.; Rudas, M.; Jakesz, R.; Dubsky, P.; Fitzal, F.; Singer, C.F.; Dietze, O.; Greil, R.; Jelen, A.; Sevelda, P.; et al. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2011, 17, 6012–6020.
- Sestak, I.; Martín, M.; Dubsky, P.; Kronenwett, R.; Rojo, F.; Cuzick, J.; Filipits, M.; Ruiz, A.; Gradishar, W.; Soliman, H.; et al. Prediction of chemotherapy benefit by EndoPredict in patients with breast cancer who received adjuvant endocrine therapy plus chemotherapy or endocrine therapy alone. Breast Cancer Res. Treat. 2019, 176, 377–386.
- Dubsky, P.C.; Singer, C.F.; Egle, D.; Wette, V.; Petru, E.; Balic, M.; Pichler, A.; Greil, R.; Petzer, A.L.; Bago-Horvath, Z.; et al. The EndoPredict score predicts response to neoadjuvant chemotherapy and neoendocrine therapy in hormone receptor-positive, human epidermal growth factor receptor 2-negative breast cancer patients from the ABCSG-34 trial. Eur. J. Cancer 2020, 134, 99–106.
- Soliman, H.; Wagner, S.; Flake, D.D., II; Robson, M.; Schwartzberg, L.; Sharma, P.; Magliocco, A.; Kronenwett, R.; Lancaster, J.M.; Lanchbury, J.S.; et al. Evaluation of the 12-Gene Molecular Score and the 21-Gene Recurrence Score as Predictors of Response to Neo-adjuvant Chemotherapy in Estrogen Receptor-Positive, HER2-Negative Breast Cancer. Ann. Surg. Oncol. 2020, 27, 765–771.
- Bertucci, F.; Finetti, P.; Viens, P.; Birnbaum, D. EndoPredict predicts for the response to neoadjuvant chemotherapy in ER-positive, HER2-negative breast cancer. Cancer Lett. 2014, 355, 70–75.
- Mazo, C.; Barron, S.; Mooney, C.; Gallagher, W.M. Multi-Gene Prognostic Signatures and Prediction of Pathological Complete Response to Neoadjuvant Chemotherapy in ER-positive, HER2-negative Breast Cancer Patients. Cancers 2020, 12, 1133.
- Andre, F.; Ismaila, N.; Henry, N.L.; Somerfield, M.R.; Bast, R.C.; Barlow, W.; Collyar, D.E.; Hammond, M.E.; Kuderer, N.M.; Liu, M.C.; et al. Use of Biomarkers to Guide Decisions on Adjuvant Systemic Therapy for Women with Early-Stage Invasive Breast Cancer: ASCO Clinical Practice Guideline Update—Integration of Results From TAILORx. J. Clin. Oncol. 2019, 37, 1956–1964.
- Sparano, J.A.; Gray, R.J.; Makower, D.F.; Pritchard, K.I.; Albain, K.S.; Hayes, D.F.; Geyer, C.E.; Dees, E.C.; Goetz, M.P.; Olson, J.A.; et al. Adjuvant Chemotherapy Guided by a 21-Gene Expression Assay in Breast Cancer. N. Engl. J. Med. 2018, 379, 111–121.
- Cardoso, F.; van’t Veer, L.J.; Bogaerts, J.; Slaets, L.; Viale, G.; Delaloge, S.; Pierga, J.Y.; Brain, E.; Causeret, S.; DeLorenzi, M.; et al. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. N. Engl. J. Med. 2016, 375, 717–729.
- Gnant, M.; Filipits, M.; Greil, R.; Stoeger, H.; Rudas, M.; Bago-Horvath, Z.; Mlineritsch, B.; Kwasny, W.; Knauer, M.; Singer, C.; et al. Predicting distant recurrence in receptor-positive breast cancer patients with limited clinicopathological risk: Using the PAM50 Risk of Recurrence score in 1478 postmenopausal patients of the ABCSG-8 trial treated with adjuvant endocrine therapy alone. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2014, 25, 339–345.
- Prat, A.; Galván, P.; Jimenez, B.; Buckingham, W.; Jeiranian, H.A.; Schaper, C.; Vidal, M.; Álvarez, M.; Díaz, S.; Ellis, C.; et al. Prediction of Response to Neoadjuvant Chemotherapy Using Core Needle Biopsy Samples with the Prosigna Assay. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 560–566.
- Schmid, P.; Cortes, J.; Dent, R.; Pusztai, L.; McArthur, H.; Kümmel, S.; Bergh, J.; Denkert, C.; Park, Y.H.; Hui, R.; et al. Event-free Survival with Pembrolizumab in Early Triple-Negative Breast Cancer. N. Engl. J. Med. 2022, 386, 556–567.
