Ischemic stroke (IS), the leading cause of death and disability worldwide, is caused by many modifiable and non-modifiable risk factors. This complex disease is also known for its multiple etiologies with moderate heritability. Polygenic risk scores (PRSs), which have been used to establish a common genetic basis for IS, may contribute to IS risk stratification for disease/outcome prediction and personalized management. Statistical modeling and machine learning algorithms have contributed significantly to this field. For instance, multiple algorithms have been successfully applied to PRS construction and integration of genetic and non-genetic features for outcome prediction to aid in risk stratification for personalized management and prevention measures. PRS derived from variants with effect size estimated based on the summary statistics of a specific subtype shows a stronger association with the matched subtype. The disruption of the extracellular matrix and amyloidosis account for the pathogenesis of cerebral small vessel disease (CSVD). Pathway-specific PRS analyses confirm known and identify novel etiologies related to IS.
- genome-wide association study
- ischemic stroke
- stroke subtypes
- cerebral small vessel disease
- polygenic risk score
- machine learning
- electronic health records
- gene ontology
- least absolute shrinkage and selection operator (LASSO)
- survival analysis
Polygenic Nature of Ischemic Stroke
Ischemic stroke (IS) is a highly complex and heterogeneous disorder caused by multiple etiologies with moderate heritability. Monogenic forms of IS are rare. Some studies have reported 30% to 40% phenotypic variability explained by common genetic variation. All main classification methods stratify IS subtypes into five major categories: large artery atherosclerosis (LAS), cardiac embolism (CES), small artery occlusion (SVS), uncommon causes, and undetermined causes. The focus of this article is to dissect the etiology of IS through pathway analyses and highlight how statistical methods and machine learning algorithms have contributed to the integration of genetic information into risk models. A flow chart summarizing the topics covered by this review to guide the reader is presented in Figure 1. We first briefly review the genetic basis of monogenic stroke and then turn our attention to the polygenic nature of sporadic IS using polygenic risk scores (PRSs) derived from large-scale genome-wide association studies (GWAS) and meta-analyses of GWAS as a tool to establish a common genetic basis for IS. We will discuss how the polygenic risk for cardiovascular disease may also contribute to the risk for sporadic IS. We will show how PRS may augment IS subtyping and review the polygenic basis of IS subtypes, such as cardioembolic stroke, cerebral small vessel disease (CSVD), and cerebral vascular amyloidosis. Our main focus is pathway-specific PRS analyses. We will show how this approach can leverage information to confirm known and identify novel etiologies related to IS. Some of these specific PRSs may contribute to IS risk stratification for disease/outcome prediction and personalized management. Finally, we will discuss the challenges of integrating PRS into clinical decision support systems and risk stratification procedures.
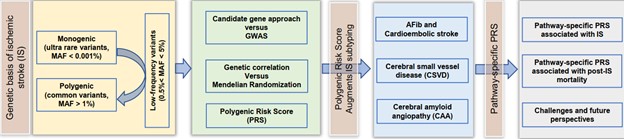
Figure 1. Flow chart summarizing the topics discussed in this review article.
- Pioneer Studies on Monogenetic Disease
Genetic studies contribute significantly to our understanding of the causality of IS and its subtypes. With reference to previous linkage studies, several distinct single-gene variants have been discovered among patients with lacunar stroke and CSVD. CSVD is a common cause of stroke and cognitive impairment in the elderly and affects small vessels of the brain, including small arteries, arterioles, capillaries, and small veins. So-called monogenic cerebrovascular diseases include: (1) cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL), which is the most prevalent monogenic CSVD and is caused by a cysteine-altering mutation in one of the 34 epidermal growth factor-like repeat (EGFr) domains of NOTCH3 gene at 19q1 1; (2) cerebral autosomal recessive arteriopathy with subcortical infarcts and leukoencephalopathy (CARASIL), which is caused by missense mutations in HTRA1, encoding a serine protease, located at 10q26.13 1; (3) Fabry disease (FD), a rare X-linked inborn error of glycosphingolipid metabolism resulting from reduced production of lysosomal α-galactosidase A (α-Gal A), resulting in the accumulation of glycosphingolipids 2 in various cellular compartments, causing structural damage and cellular dysfunction and triggering a secondary inflammatory response, resulting in progressive organ dysfunction 1; (4) retinal vasculopathy with cerebral leukodystrophy, an autosomal dominant disorder caused by C-terminal frameshift mutations in the Three Prime Repair Exonuclease 1 (TREX1) gene located at 3p21.313; (5) COL4A1/COL4A2-related angiopathies; COL4A1/A2, located at 13q34, encodes the most abundant and prevalent protein in the basement membrane of all tissues, including cerebral vasculature; type IV collagen helps the basement membrane interact with other cells, playing a role in cell migration, proliferation, differentiation, and survival; and (6) hereditary cerebral amyloid angiopathy (CAA), characterized by cerebrovascular amyloid deposition, mainly observed in leptomeningeal and cortical vessels; it can be classified based on accumulated amyloid proteins, such as amyloid β (APP), cystatin C (CST2), integral membrane protein 2B (ITM2B), prion protein, transthyretin (TTR), and others4.
Understanding the genetics of monogenic CSVD and lacunar stroke5 can lead to precise diagnosis and prognosis, aid in the development of a targeted treatment plan, and ultimately lead to an improved phenotype definition. Monogenic diseases are rare, and the causal variants have a minor allele frequency (MAF) of less than 0.005 (ultra-rare) in the stroke population. Sporadic IS, which dominates the disease population, cannot be explained by these rare inheritances despite some success in identifying common risk loci at the gene level (e.g., COL4A2 and HTRA1) by the GWAS 1, 6.
- Low-Frequency Variants Explain More Phenotypic Variation
Previously identified IS risk loci with the significant genome-wide association are enriched with low-frequency variants7. The partition of SNPs by MAF can provide deep insight into the mechanisms of heritability. If a genetic variant is associated with fitness, selection would drive one allele to low frequency8. The latter is the case even for traits without any obvious connection to fitness. The functional architecture of low-frequency variants (0.5% < MAF < 5%) highlights the strength of negative selection across coding and non-coding variants; this effect is also obvious with respect to many cardiometabolic traits9. Low-frequency variants bridge the gap between rare variants with putatively larger effect sizes and common variants with smaller effect sizes. Because the loci for cardiovascular diseases are significantly enriched for lifetime reproductive success by natural selection10 and identified IS subtype-specific loci are more likely to be low in MAF7, 11, we propose that genetic variants with lower MAF may contribute more to the phenotypic variation in IS. When we partitioned the variants by MAF ≤ 0.01, 0.05, 0.1, 0.2, or to all, PRSLAS, PRSCES, and PRSSVS derived from low-frequency common variants (0.01 < MAF < 0.05) provided the best-fit modeling for our IS cohort, suggesting that low-frequency common variants, when taken together, could contribute more to the risk for matched IS subtypes.
- Polygenic Risk Scores (PRSs) Augment IS Subtyping
PRSs derived from stroke subtypes may augment the predictive power for patients with a similar etiology. PRSs for atrial fibrillation can significantly explain cardioembolic stroke (CES) risk, independent of other clinical risk factors12.
We previously showed that PRSLAS, PRSCES, and PRSSVS, which were constructed by the variants with effect size estimated according to the MEGASTROKE IS subtypes (LAS, CES, or SVS), explained the most variance of the corresponding subtypes of IS among MEGASTROKE subtypes (larger and warmer dots for the significant level and Nagelkerke pseudo-R2, respectively, using variants from the base file with p < 0.1). To determine the robustness of this subtype-specific PRS, a synthesized group (ASL) with more LAS cases (n = 120) than SVS cases (n = 70) was created. We observed that the predictive power (R2) and significance was the highest using PRSsvs, suggesting that there is a lack of a clear boundary between LAS and SVS. However, PRSCES differentiated LAS from CES and SVS from CES (yellow arrows), suggesting that CES has a unique polygenic architecture that separates it from other subtypes. Furthermore, none of the PRSs could significantly explain the phenotypic variation of our ‘Undetermined’ subtype. In summary, some clinical IS subtypes may have distinct or shared polygenic architecture. The effect sizes from low-frequency variants estimated by the summary statistics of GWAS on clinical subtypes contribute more to the polygenic inheritance of the matched subtype.
- A Modified Paradigm of IS Risk Stratification beyond TOAST Subtyping
The primary goal of diagnostic stroke evaluation is to identify the underlying etiology so that targeted treatments can be designed and implemented to prevent a recurrence13. Several classification systems have managed to stratify stroke etiologies into discrete clinical, radiographic, and prognostic categories. Despite a decade of GWAS on IS and its subtypes, genetic evidence currently has only been considered under certain circumstances, in which prothrombotic abnormalities should be considered as a cause of stroke exclusively in patients with a history of unexplained thromboembolic events in young stroke patients who have no other explanations for their stroke14-16. There is an unmet need for the etiologic classification of strokes with multiple potential mechanisms into specific etiologic classes in the absence of evidence-based strategies, such as risk factors, family history, and medication, and to better quantify multiple competing causes in a given patient17, 18. How genetic information from GWAS contributes to this etiologic classification of strokes and may assist in identifying the etiology of strokes of unknown origin, referred to as cryptogenic strokes, is still unclear. Mechanism-targeted treatments are not available for cryptogenic strokes, which represent 25% to 30% of IS, increasing the likelihood of have recurrent events. The quality of etiologic classification depends on the ability to generate homogenous subtypes with discrete outcomes (discriminative validity) and the clarity of classification rules to ensure utility in different settings with different investigators (reliability)13. It is necessary to further categorize IS using more homogenous groups stratified by risk factors, including PRS, and refine the current diagnostic system for subtyping. Whether PRS may augment the newer clinical classification systems (e.g., ASCO and CCS) should be determined, as these newer schemes may better stratify the stroke etiology, at least in some patients. Based on the pioneer studies by consortia19, 20 and our PRS modeling and that of others12, 21, we propose a modified paradigm of IS risk stratification beyond TOAST subtyping to incorporate genetic information into the existing ideological classification system.
- Improved Predictability of Pathway-Specific PRS for Post-IS Mortality Using an Integrated Cox Proportional Hazards Model
Improved predictability can be achieved by better interrogating the data and by using methodologies that are carefully aligned with the data characteristics. Owing to the hierarchical nature of GO biological process terms, multicollinearity of PRSs is common. There are also extensive correlations within or between PRSs and clinical risk factors. All these factors can inflate the regression coefficients of predictive variables in the multivariate regression model. An L1 penalization technique (LASSO regression) can handle this situation by forcing some regression coefficient estimates to be exactly zero, thus achieving variable selection while shrinking the remaining coefficients toward zero to avoid the overfitting and overestimation caused by data-driven model selection. The least absolute shrinkage and selection operator (LASSO) method22 in the multivariate Coxph model was applied for feature selection of prognostic pathway-specific PRSs23. A prediction model including an additional 16 disease-associated pathway-specific PRSs outperformed the base model (8 clinical risk factors), as demonstrated by a higher concordance index (0.754, 95% CI: 0.693–0.814 versus 0.729, 95% CI: 0.676–0.782, respectively) in the holdout sample (p < 0.001 for the median improvement). Compared to the base model, the integrated PRS prediction model differentiated not only the high-risk from the intermediate-risk (p = 0.006) but also the intermediate-risk from the low-risk (p = 0.001). The PRS derived from GO negative regulation of endothelial apoptotic pathway was the independent predictor for 3-year post-IS mortality (HR = 1.203)23.
Conclusion
In this paper, we discussed the polygenic nature of IS and emphasized the role of PRS in risk stratification for disease/outcome prediction and personalized management of IS. Polygenic risk for cardiovascular disease may also account for the risk of sporadic IS. PRS may augment IS subtyping. We introduced a pathway-specific PRS analysis and demonstrated its utility in confirming known and identifying novel etiologies of IS. Some of these specific PRSs (e.g., derived from the endothelial cell apoptosis pathway) individually contribute to post-IS mortality, and together with clinical risk factors, better predicted post-IS mortality. Statistical models and machine learning algorithms have contributed significantly to the advancement in this field and will continue to drive innovation for genome-based healthcare decision making striving toward innovation, equity, and transparency.
Reference:
- Germain DP. Fabry disease. Orphanet J Rare Dis 2010;5:30.
- Zhou C, Huang J, Cui G, Zeng H, Wang DW, Zhou Q. Identification of a novel loss-of-function mutation of the GLA gene in a Chinese Han family with Fabry disease. BMC Med Genet 2018;19:219.
- Richards A, van den Maagdenberg AM, Jen JC, et al. C-terminal truncations in human 3'-5' DNA exonuclease TREX1 cause autosomal dominant retinal vasculopathy with cerebral leukodystrophy. Nat Genet 2007;39:1068-1070.
- Yamada M. Cerebral amyloid angiopathy: an overview. Neuropathology 2000;20:8-22.
- Traylor M, Persyn E, Tomppo L, et al. Genetic basis of lacunar stroke: a pooled analysis of individual patient data and genome-wide association studies. Lancet Neurol 2021;20:351-361.
- Li J, Abedi V, Regeneron Genetic C, Zand R, Griessenauer CJ. Replication of Top Loci From COL4A1/2 Associated With White Matter Hyperintensity Burden in Patients With Ischemic Stroke. Stroke 2020;51:3751-3755.
- Malik R, Traylor M, Pulit SL, et al. Low-frequency and common genetic variation in ischemic stroke: The METASTROKE collaboration. Neurology 2016;86:1217-1226.
- Wray NR, Yang J, Hayes BJ, Price AL, Goddard ME, Visscher PM. Pitfalls of predicting complex traits from SNPs. Nat Rev Genet 2013;14:507-515.
- Gazal S, Loh PR, Finucane HK, et al. Functional architecture of low-frequency variants highlights the strength of negative selection across coding and non-coding annotations. Nat Genet 2018;50:1600-1607.
- Byars SG, Huang QQ, Gray LA, et al. Genetic loci associated with coronary artery disease harbor evidence of selection and antagonistic pleiotropy. PLoS Genet 2017;13:e1006328.
- Li J, Chaudhary DP, Khan A, et al. Polygenic Risk Scores Augment Stroke Subtyping. Neurol Genet 2021;7:e560.
- Pulit SL, Weng LC, McArdle PF, et al. Atrial fibrillation genetic risk differentiates cardioembolic stroke from other stroke subtypes. Neurol Genet 2018;4:e293.
- Arsava EM, Helenius J, Avery R, et al. Assessment of the Predictive Validity of Etiologic Stroke Classification. JAMA Neurol 2017;74:419-426.
- Bushnell CD, Goldstein LB. Diagnostic testing for coagulopathies in patients with ischemic stroke. Stroke 2000;31:3067-3078.
- Bushnell C, Siddiqi Z, Morgenlander JC, Goldstein LB. Use of specialized coagulation testing in the evaluation of patients with acute ischemic stroke. Neurology 2001;56:624-627.
- Waddy SP. Disorders of coagulation in stroke. Semin Neurol 2006;26:57-64.
- Ay H, Furie KL, Singhal A, Smith WS, Sorensen AG, Koroshetz WJ. An evidence-based causative classification system for acute ischemic stroke. Ann Neurol 2005;58:688-697.
- Ay H, Benner T, Arsava EM, et al. A computerized algorithm for etiologic classification of ischemic stroke: the Causative Classification of Stroke System. Stroke 2007;38:2979-2984.
- Malik R, Chauhan G, Traylor M, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet 2018;50:524-537.
- Network NSG, International Stroke Genetics C. Loci associated with ischaemic stroke and its subtypes (SiGN): a genome-wide association study. Lancet Neurol 2016;15:174-184.
- Abraham G, Malik R, Yonova-Doing E, et al. Genomic risk score offers predictive performance comparable to clinical risk factors for ischaemic stroke. Nat Commun 2019;10:5819.
- Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med 1997;16:385-395.
- Li J, Abedi V, Zand R. Predicting mortality among ischemic stroke patients using pathways-derived polygenic risk scores. Sci Rep 2022.
This entry is adapted from the peer-reviewed paper 10.3390/jcm11205980
