Cellulolytic enzyme synergism is essential for deconstructing cellulose substrates in the biorefinery sector. Therefore, research on cellulolytic enzyme synergistic interactions has attracted extensive interest worldwide. Cellulases, consisting of endo-glucanases, exoglucanases and beta-glucosidases, were thought to be the main enzymes that degrade or modify cellulosic substrates. However, an accumulation of information on the modes of action and substrate hydrolysis mechanisms of expansins, swollenis (SWOs) and lytic polysaccharide mono-oxygenases (LPMOs) revealed that they significantly improve cellulolytic enzyme synergism. An up-to-date model of cellulose deconstruction, including these novel proteins, is required for effective cellulose degradation and advancement of cellulose-based biorefineries.
- cellulase
- cellulose
- expansin
- LPMO
- synergy
- swollenins
1. Introduction
Lignocellulosic feedstocks have enormous economic potential as a source of value-added products (VAPs) and biofuels [1][2][3]. Cellulose is the most abundant polymer on earth and a predominant component of lignocellulosic biomass. It is a source of fermentable sugars (glucose and cello-oligosaccharides) that serve as precursors for VAPs. Cellulose is a linear polymer that consists of β-D-glucose molecules linked by glycosidic bonds, sourced from plant material (e.g., wood pulp, cotton, or cereals such as wheat, sugarcane and rice bagasse) [1][4][5][6]. Many cellulose chains bundle via hydrogen bonding to constitute cellulose microfibrils, which consist of crystalline regions intersected by amorphous regions.
It is established that glycoside hydrolases (GHs) from different families act synergistically to produce fermentable sugars from the lignocellulose biomass [3][7][8][9][10]. The cellulolytic enzyme synergism is based on the simultaneous use of the cellobiohydrolases; CBHI (EC 3.2.1.176) and CBHII (EC 3.2.1.91) which attack the biomass from the reducing and non-reducing ends of cellulose chains to produce cellobiose, respectively, endoglucanase (EG, EC 3.2.1.4) which randomly cleave the amorphous regions of the to produce oligosaccharides with a DP between 2 and 10 or create new chain ends in the bulk of biomass, and β-glucosidase (β-gl, EC 3.2.1.21) which degrade oligosaccharides to glucose [9]. Figure 1 shows the well-established mode of action of cellulases during the synergistic hydrolysis of cellulose.
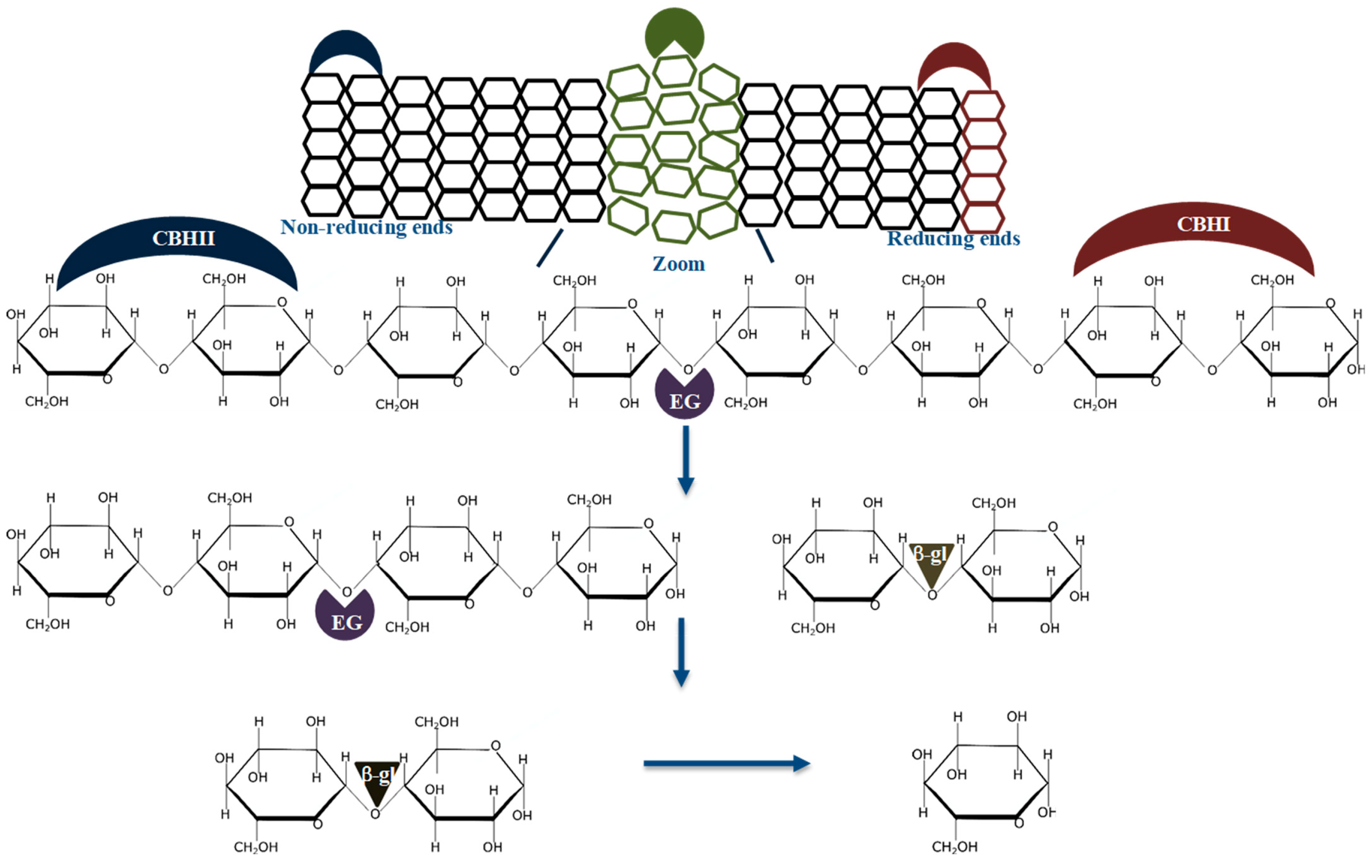
The cellulolytic enzyme synergism can be grouped into two classes; (1) synergism between non-catalytic and catalytic cellulolytic enzymes (e.g., expansins and CBHs), and (2) synergism between catalytic cellulolytic enzymes; i.e., Exo-Exo, Exo-Endo, Exo-Endo-β-gl or cellulase and lytic polysaccharide monooxygenase (LPMO) synergy [11][12][13][14][15][16]. A few studies have documented Endo-Endo synergy, even though EGs constitute most diverse cellulase group, classified under GH families 5, 6, 7, 8, 9, 10, 12, 44, 45, 48, 51, 74 and not classified sequences (NC) (http://www.cazy.org/Glycoside-Hydrolases.html). Also, it is well documented that CBHs from different GH families that hydrolyze cellulose on the same cleavage sites do not synergize together, but compete for occupancy on the same binding sites [17][12].
Two other enzyme systems play a significant role in cellulose saccharification, these being LPMOs and non-hydrolytic enzymes (expansins and swollenins (SWOs)). According to the CAZy database, the enzymes classified under CBM33 were later assigned to auxiliary activities family 10 (AA10), while those in GH61 were assigned to AA9 [http://www.cazy.org/Auxiliary-Activities.html, [18]. The LPMOs in family AA9 and AA10 are copper-dependent enzymes that cleave the glycosidic bonds of crystalline cellulose regions by oxidizing the C1 (EC 1.14.99.54) or C4 (EC 1.14.99.56) and C1/C4 of the glucose residues ([18][19], http://www.cazy.org/Auxiliary-Activities.html]. The LPMOs in family AA16 are newly classified cellulose-specific enzymes with a C1 regiospecificity [20]. The catalytic domain of LPMOs can also be attached to CBMs by a flexible linker [19][21].
Expansins and expansin-like proteins are nonhydrolytic or nonoxidative enzymes that can bind to the crystalline cellulose and weaken the degree of crystallinity of the polymer by breaking the inter- or intramolecular hydrogen bonding network [22][23]. Kim et al. [24] demonstrated that Bacillus subtilis derived expansin sequences were homologous to maize plant expansins, and their 3D structures were similar. The role of expansins in plants is well established, and plant-derived expansins are classified into two groups, α- and β-expansins [4]. The α- and β-expansins only share 20% amino acid sequence similarity. Interestingly, expansins generally have about 30% sequence similarity to endoglucanases classified under GH45 [4]. Several studies have shown that the expansins synergize with cellulases that hydrolyze crystalline microfibrils of cellulose [4][22][23][25]. Fungi also possess another nonhydrolytic protein called swollenins, similar to the expansins [25]. Swollenins modify the chemical and structural features of microcrystalline cellulose resulting in a reduced degree of crystallinity, which allow the cellulases to hydrolyze cellulose effectively. Additionally, studies have demonstrated that cellulases and swollenins synergize to improve the saccharification of cellulose [12][25]. Herein, hydrolytic cellulolytic enzyme to non-hydrolytic enzyme synergism and the degrees of synergism (DS) thresholds to develop a model that could assist in the improvement of cellulose hydrolysis by cocktails will be explained.
Lack of investigation of the DS during synergism is attributed to a focus on hydrolysis yields by researchers. It appears that some enzyme combinations do not display a positive DS, yet these combinations demonstrate higher yields in terms of soluble sugar production compared to when the individual enzymes are used to hydrolyze the substrate. However, it is proposed that investigating synergism between enzymes should not only be limited to yield improvement but should also include DS. This would enable researchers and companies to design better enzyme cocktails for specific feedstocks. It will be addressed how can it could be achieved that higher yields and DS by reevaluating the established synergisms and setting the DS-threshold benchmarks, which can lead to higher yields.
2. Defining DS and Its Impact on Substrate Hydrolysis
It is well accepted that a DS of 1 arbitrary unit (AU) demonstrates no synergism between the enzymes used to formulate a specific enzyme cocktail [9][10][17][26]. These studies suggest that the enzymes do not cooperate during substrate hydrolysis leading to no improvement in the rate of substrate deconstruction compared to when the mono-components act individually. For example, endoglucanase and exoglucanase enzymes do not synergize and generally result in a DS of 1 AU, during the hydrolysis of the model amorphous cellulose, carboxyl-methylcellulose (CMC), because of their uncomplimentary modes of action on the substrate. A value below 1 AU shows that there is no synergism between the enzymes during the hydrolysis of the substrate. This observation suggests that one or more of the enzymes cannot hydrolyze the substrate efficiently due to competitive binding of two or more enzymes on the same substrate cleavage site. Instances which resulted in a DS below 1 AU have been documented between CBHII and EG, processive EGs from GH9. In all these cases, substrate hydrolysis was impeded, and product formation significantly reduced. Generally, DS values above one are achieved when the enzymes in the cocktails hydrolyze different cleaving sites of the substrate, e.g., CBHI, CBHII, EG and β-gl degrade substrates with different cleaving sites, resulting in significantly higher product formation (Figure 1).
3. Synergism between catalytic enzymes
It was hypothesized that to formulate effective enzyme cocktails researchers should not only focus on the yield of soluble sugars as conventional synergy studies have done for the past several decades but should focus on both the yield of soluble sugars and the DS between enzymes. This approach will generate holistic information about the reaction with regards to the interactions between the enzymes in the cocktails, as well as the products formed. Hence, it is attempted to establish the DS threshold which results in high yields of soluble sugars. A detailed discussion of various cellulolytic enzyme synergy studies that were conducted with different enzyme combinations are presented below and summarized in Table 1.
| Type Synergy | Cellulolytic Enzymes | Cellulose Substrate | Conversion Yield/Activity Increase | Recorded/Predicted # DS Values | DS-Threshold * | Reference |
|---|---|---|---|---|---|---|
| Non-catalytic and catalytic active cellulolytic enzyme synergy | Expansins: Cel7A or Cel7B | model cellulose II films |
<5-fold | N/A | DS value > 1 | [31] |
| Bpexpansins and celluclast™ Bp- or Cm-expansins: celluclast™ |
Avicel PASC Filter paper |
<2-fold <1-fold <5-fold |
2.5 1.3 7.3 |
[18] [19] |
||
| Bsexpansin: cellulase | Filter paper | <2-fold | 2.5 | [42] | ||
| Xcexpansin: Accellerase 1500 | Filter paper | 36% | 1.4 | [46] | ||
| TrSWO: CHI or EGII | Valonia cell walls | N/A | N/A | [33] $ | ||
| Po-SWO: cellulases | Avicel | <2-fold | 2.2 | [17] | ||
| Processive Cellulolytic enzyme synergism (Exo-Exo &Exo-processive Endo a) | HjCel6A: HjCel7A HiCel6A: HiCel7A Cel6A: Cel7A |
Avicel Bacterial cellulose Mercerized Avicel |
<3-fold 90% <1.2-fold |
2.3 3 1.4 |
DS value > 1.2 | [20] [44] [15] |
| Cel9A: Cel48A EG: CBH CcCel9A: CcCel48 HiCel6A: HiCel7A |
Filter paper Filter paper Filter paper Bacterial cellulose |
17% <3-fold <1.2-fold 30% |
1.7 2.6 1.7 4.5 |
[47] [48] [49] [44] |
||
| Endo-Endo synergism | CcCel9A: CcCel9B CelZ: CelY |
CMC CMC |
2-fold 2-fold |
1.1 1.8 |
ND | [49] [50] |
| Endo-Beta-glucosidase | CcCel9A: BlgA CgEG1: CgBlg1 CgEG1: CgBlg1 EG: Bgl EG: Bgl c |
Filter paper CMC Sigma-cell Filter paper DMOS-SCB |
<1.8-fold <90% <80% 1.75-fold 3-fold |
2.1 9 2.8 2.0 3 |
DS value > 1.3 | [49] [51] [51] [51] [51] |
| Endo-Exo synergy b | TrCel7A: TrCel5A Cel6: Cel5A ThCel7B: ThCel7A HiCel7A: HiCel45A HiCel6A:HiCel45A HiCel6A:HiCel7A: HiCel45A TrEGII: TrCBHI: TrCBHII CBHI: EGII: CBHI CcCel9A:CcCel9BCcCel48A TrCel7A: TrCel7B |
Bacterial cellulose Cellulose-III Filter paer Bacterial cellulose Bacterial cellulose Bacterial cellulose Avicel Cellulose with DP3000 Avicel Filter paper steam-pretreated spruce |
N/A 5% 92% 25% 15% 90% 25% N/A 27% <3-fold <2-fold |
1.7 2.16 1.6 2.6 2.1 2.0 2 2.6 1.6 2.6 1.8 |
DS value > 2 | [16] [52] [14] [44] [44] [44] [53] [5] [54] [49] [55] |
| Cellulases and LPMO synergism | AaAA16: CBHI Celluclast: mgLPMO10 TtMO9E: Cel6A Celluclast: CelS2 MtLPMO9L: CBHII TrCel7A: TrCel6A: TrCel7B: TtAA9 MtEG5A: MtEG7A: MtLPMO9 |
PASC Avicel PASC Filter paper PASC PASC PASC |
<1.8-fold 36% 2-fold 4-fold 3-fold 2.8-fold <2.5-fold |
1.8 1.4 1.9 4 3 3 2.6 |
DS values > 2 | [25] [26] [37] [23] [56] [29] [57] |
Bp: Bacillus pumilus, Bs: Bacillus subtilis, Xc: Xanthomonas campestris, Cm: Micromonospora aurantiaca ATCC 27029, Tr-SWO: Trichoderma reesei-swollenin, Th: Trichoderma harzianum, Tl: Trichoderma longibrachiatum, Po: Penicillium oxalicum, Hj: Hypocrea jecorina, Hi: Humicola insolens, Cc: Clostridium cellulosi, Ba: Bacillus amyloliquefaciens mg: derived from metagenome, Aa: Aspergillus aculeatus, Mt: Myceliophthora thermophila, Tt: Thermothelomyces thermophilus. # Predicted DS values were estimated directly from published data. * Theoretical DS value thresholds which generally result in higher degrees of synergy. $ Solubility studies which were equivalent to synergistic activity. a Conversional exo–exo synergy can be substituted with processive EG in a new approach of exo-processive endo. b Endo–exo synergy is generally supplemented with β-gl to prevent product inhibition. c The enzymes were immobilized on synthetized Fe3O4 nanoparticles.
3.1. Synergism between processive cellulolytic enzymes; Exo-Exo or Exo-processive Endo synergy
There is strong evidence that supports the hypothesis that some backbone-cleaving enzymes always interact synergistically, resulting in a DS that is higher than 1 AU and a soluble sugar yield which is higher than that produced by the sum of the individual enzymes during the hydrolysis of biomass [9]. For instance, cellulase enzymes that cleave the microcrystalline cellulose, such as Cel6A and Cel7A (exo-exo synergy) or Cel6A, Cel7A and EG (exo-endo synergy) generally synergize with a DS that is higher than 1 AU [9][17][15]. Exo-exo synergy between cellulases has been demonstrated by Boisset et al. [27] using bacterial microcrystalline cellulose (BMC), while other researchers from the laboratory have used Avicel [phosphoric acid swollen cellulose (PASC) or NaOH-treated Avicel also called regenerated amorphous cellulose (RAC)] as a suitable substrate [10][17][28]. Badino et al. [15] showed that the DS between GH7 (Cel7A) and GH6 (Cel6A) enzymes sourced from Hypocrea jecorina was above 2.2 AU, while the glucose yield produced by synergistic actions was 4-fold higher than that which was produced by individual enzymes. A study by Boisset et al. [29] also found similar results using Humicola insolens GH7 Cel7A and GH6 Cel6A which showed a DS of about 4 AU between enzyme combinations and converted more than 50% of the bacterial microcrystalline cellulose compared to 15% by Cel6A and 28% by Cel7A alone. Based on the literature and the experience, the fungal sourced Cel7A and Cel6A synergism generally display a DS above 2 AU and will always result in a higher yield of soluble sugars than individual enzymes.
In some bacterial systems such as Clostridium phytofermentans ISDg, there is a different cellulase system that degrades the crystalline regions of cellulose [30]. The CBH enzymes employed by C. phytofermentans ISDg are Cel48 and Cel9. Zhang and co-workers suggested that Cel48 attaches to the substrate from the reducing end, while Cel9 (processive EG) attaches from the non-reducing end. In addition, CBHs that belong to GH 48, such as Cel48, usually contain a family 3 carbohydrate-binding domain (CBM) [30]. Anaerocellum thermophilum is another bacterium which has demonstrated that the GH48 and GH9 enzyme system is generally employed to hydrolyse microcrystalline cellulose via exo-exo synergy [31]. Additionally, Irwin and co-workers showed that there was higher synergism between Cel48A and Cel9A sourced from Thermobifida fusca compared to other enzymes from different GH families like GH5 and GH6 [30]. Cel9 from Bacillus licheniformis was shown to have a CBM3c linked to the catalytic domain and displayed the highest activity on PASC, BC and filter paper. Cel9 (GH 9) from Clostridium cellulosi possesses five CBMs, namely; CBM3c, three CBMX2s and CBM3b [48]. The exo-exo synergy between the Cel48 and Cel9A on the filter paper displayed a DS value of 1.5 AU [32]. Another study similarly reported a high DS of about 1.5 AU for Cel48 (GH48) and Cel9 (GH9), while the soluble sugars produced from Avicel was 4-fold higher than those produced by the individual enzymes [30]. These observations support the hypothesis that there are two or more distinct exo-cellulase systems that result in high DS and yields of soluble sugars; (1) one employed by both bacterial and fungal cellulolytic systems (mostly Cel7A and Cel6A), and (2) another unique to the bacterial cellulolytic system (Cel 48 and Cel 9A).
3.2. Exo-Endo and/or β-glucosidase synergy (cellulolytic enzyme cocktail)
Based on both theoretical and empirical evidence, CBHs, EGs and β-gls can synergize during the hydrolysis of biomass (Figure 1). Boisset et al. [33] demonstrated that CBHI (Cel7A), CBHII (Cel6A) and EG (Cel45A) sourced from Humicola insolens displayed synergism during the hydrolysis of bacterial cellulose ribbons (micro-crystalline cellulose). The composition of Cel6A and Cel7A were varied between 0 and 100%, while Cel45A was fixed at 1.2%. The cocktail composed of the three enzymes converted more than 60% of the bacterial cellulose ribbons and displayed a DS of about 4.5 AU, while the combination of Cel6A and Cel7A converted only 30% and had a DS of 2 AU [29]. Some of the commercial cocktails such as Celluclast® 1.5L consist of Trichoderma reesei cellulases in the following combinations 55% Cel7A, 10% Cel6A, 10% Cel7B, 10%, Cel5A and 1% β-gl . The highest biomass conversion that the authors could achieve was about 50% yield, only when they added an extra 10% β-gl – but their experimental design did not factor in the DS, which shows the molecular level interactions between the enzymes. Addition of 50 U/g substrate of a GH12 EG sourced from Gloeophyllum trabeum with 10 U/g substrate of Celluclast® 1.5L resulted in an increased synergistic effect of about 14.5%, 16.1%, 29.0% and 13.4% on filter paper, hydrogen peroxide–acetic acid-pretreated pine, corn stover and rice straw, respectively [34].
The above information relates to the fungal cellulolytic enzyme cocktails; however, it is clear that exoglucanase systems of bacterial organisms are different to those of fungal systems (as it was proposed in the Exo-Exo synergy section). For instance, Cel6A, Cel48A, Cel6B and Cel9A sourced from Cellulomonas fimi were all processive on PASC, Avicel and crystalline celluloses Iα or IIII from green algae [18]. It is important to note that Cel6B and Cel9A are classified as endoglucanases, but Uchiyama et al. [18] demonstrated that these enzymes are arguably highly processive and used high-speed atomic force microscopy (HS-AFM) to show that they were able to move on crystalline cellulose IIII. Interestingly, CfCel6B and CfCel5B demonstrated a DS of 2.16 AU and an improved yield of soluble sugar at a ratio of 3:1 when the enzyme loading was at 2.5 mg/g cellulose concentration [35]. However, when the enzyme concentration of CfCel6B and CfCel5B was increased to 10 mg/g cellulose, the DS values were approximately decreased by 1 AU for all combinations; 3:1, 1:1 and 1:3. The processive and non-processive EGs from GH family 9 and a CBHI from GH family 48 sourced from C. cellulosi CS-4-4 displayed synergy on filter paper when they were supplemented with β-gl from Caldicellulosiruptor sp [34]. Various molar ratios were tested in this study and the best combination for the formulated cellulose cocktail was 25:25:10:18 for CcCel9A: CcCel9B: CcCel48A: BlgA, respectively, which resulted in a DS of 2.6 and 1.86 mg/ml of glucose release. The literature contains more information regarding fungal cellulolytic enzyme cocktails, but so far, only a few bacterial cellulolytic enzyme cocktails have been recorded. These suggest that there is still a lot that needs to be done with regard to bacterial cellulolytic enzymes, to establish if their synergy can be superior to those of fungi or even commercial cocktails. It is propose that a DS-threshold above 1.5 AU can result in higher yields of soluble sugars for bacterial cellulolytic cocktails, but more studies are required to establish this claim.
4. Cellulase and LPMO synergism
Synergism between an LPMO and CBHI was demonstrated by core expression of the genes that encode for these proteins in the fungus Penicillium funiculosum that produced the LPMO naturally and five engineered strains [16]. It was evident that the secretome from the engineered strains (referred to as PfOAO1 and PfOA3) contained higher Avicelase activity, filter paper activity and production of hydrogen-peroxide, which suggest the presence of cellulases and an LPMO. In addition, the use of PfOAO1 and PfOA3 for the saccharification of acid pretreated wheat straw increased conversion to 80% and 75%, respectively. These findings suggest that overexpression of CBHI and LPMO in PfOAO1 and PfOA3 increased synergy between these enzymes. Interestingly, the individual application of three LPMOs from Thermoascus aurantiacus (TaAA9A), Lentinus similis (LsAA9A) and Thielavia terrestris (TtAA9E) on Avicel and PASC resulted in no release of detectable reducing sugars according to Tokin et al [36]. In contrast, the LPMOs synergized with TrCel7A and TrCel6A on Avicel and PASC, except for TrCel7A and TtAA9E which displayed anti-synergy. The binary synergy between TrCel6A and TtAA9E resulted in the highest DS of about 2.5 and 2 AU during PASC and Avicel degradation, respectively. The high synergism between these enzymes can be explained by the fact that TtAA9E cleaves cellulose substrates by oxidizing the C1 of sugar moieties, creating new binding sites for TrCel6A. TrCel6A also synergized with TaAA9A or LsAA9A as their DS values were 2.0 and 1.8 AU on PASC, and 1.5 and 1.3 AU on Avicel [36]. The CBHI and TaAA9A or LsAA9A displayed a DS of 2.25 and 1.4 AU on PASC or 1.8 and 1.5 AU, respectively [36]. EG (Cel7B) and MtLPMO9A synergy improved hydrolysis of Avicel, bacterial cellulose and sugarcane bagasse (SCB), which resulted in higher glucose production in the presence of Anβ-gl. The DS values between cellulases (TrCel7A, TrCel6A and TrCel7B) and MtLPMO9A during the degradation of Avicel, BC and SCB were 2.8, 2.5 and 2.6 AU, respectively. We propose that DS values greater than 1.5 AU should be considered as a threshold which result in higher yields of soluble sugars during synergism between the cellulases and LPMOs from AA9 (LPMO9A) during the degradation of amorphous substrates such as PASC. However, DS values greater than 2 AU between cellulases and LPMO9A should be considered as a threshold which results in the improved degradation of crystalline substrates like Avicel.
5. Conclusion
Cellulolytic enzyme synergism is essential for the deconstruction or modification of cellulose in the bio-refinery sector. Hence, research on synergistic interactions has attracted extensive interest from all over the world over the past few decades. Cellulases are the main enzymes that degrade or modify cellulose substrates; however, expansins, SWO and LPMO demonstrate different modes of degradation action on the cellulose substrates. The LPMOs and cellulases or expansins/swollenins and cellulases synergism results in an enhanced conversion rate of the cellulose substrates to soluble sugars, aldoronic and gem-diol acids. Interestingly, to date, there have been no studies that have investigated the synergy between expansins/swollenins and cellulolytic LPMOs. This suggests that knowledge gaps exist in how expansins or swollenins and LPMOs (AA9, AA10 and AA16) interact during biomass deconstruction. Perhaps the enzyme cocktail(s) containing the expansins or swollenins, cellulases and LPMOs could be superior to the currently available commercial cellulolytic enzyme cocktails. However, it is vital to understand different forms of synergy between the enzymes before formulating enzyme cocktails containing these proteins. Lastly, the theorized DS value thresholds can be applied to understand the level of synergism between cellulolytic enzymes during the formulation of enzyme cocktails.
This entry is adapted from the peer-reviewed paper 10.3390/catal11111343
References
- McFarlane, H.E.; Doring, A.; Persson, S. The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 2014, 65, 69–94.
- Coseri, S. Cellulose: To depolymerize… or not to? Biotechnol. Adv. 2017, 35, 251–266.
- Malgas, S.; Rose, S.H.; van Zyl, W.H.; Pletschke, B.I. Enzymatic hydrolysis of softwood derived paper sludge by an in vitro recombinant cellulase cocktail for the production of fermentable sugars. Catalysts 2020, 10, 775.
- Cosgrove, D.J. Loosening of plant cell walls by expansins. Nature 2000, 407, 321–326.
- Zhang, X.-Z.; Zhang, Y.-H.P. Cellulases: Characteristics, sources, production, and applications. In Bioprocessing Technologies in Biorefinery for Sustainable Production of Fuels, Chemicals, and Polymers; Yang, S.-T., El-Enshasy, H.A., Thongchul, N., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2013.
- Jiang, F.; Ma, L.; Cai, R.; Ma, Q.; Guo, G.; Du, L.; Xiao, D. Efficient crude multi-enzyme produced by Trichoderma reesei using corncob for hydrolysis of lignocellulose. 3 Biotech. 2017, 7, 339.
- Kuhad, R.C.; Deswal, D.; Sharma, S.; Bhattacharya, A.; Jain, K.K.; Kaur, A.; Pletschke, B.I.; Singh, A.; Karp, M. Revisiting cellulase production and redefining current strategies based on major challenges. Renew. Sustain. Energy Rev. 2016, 55, 249–272.
- Thoresen, M.; Malgas, S.; Mafa, M.S.; Pletschke, B.I. Revisiting the phenomenon of cellulase action: Not all endo-and exo-cellulase interactions are synergistic. Catalysts 2021, 11, 170.
- Segato, F.; Dias, B.; Bertoa, G.L.; de Oliveira, D.M.; De Souza, F.H.M.; Citadini, A.P.; Murakami, M.T.; Damásio, A.R.L.; Squina, F.M.; Polikarpova, I. Cloning, heterologous expression and biochemical characterization of a non-specific endoglucanase family 12 from Aspergillus terreus NIH2624. Biochim. Biophys. Acta 2017, 1865, 395–403.
- Jalak, J.; Kurasin, M.; Teugjas, H.; Valjamae, P. Endo-exo synergism in cellulose hydrolysis revisited. J. Biol. Chem. 2012, 287, 28802–28815.
- Badino, S.F.; Christensen, S.J.; Kari, J.; Windahl, M.S.; Hvidt, S.; Borch, K.; Westh, P. Exo-exo synergy between Cel6A and Cel7A from Hypocrea jecorina: Role of carbohydrate binding module and the endolytic character of the enzymes. Biotechnol. Bioeng. 2017, 114, 1639–1647.
- Ogunyewo, O.A.; Randhawa, A.; Gupta, M.; Kaladhar, V.C.; Verma, P.K.; Yazdania, S.S. Synergistic Action of a Lytic Polysaccharide Monooxygenase and a Cellobiohydrolase from Penicillium funiculosum in Cellulose Saccharification under High-Level Substrate Loading. Appl. Environ. Microbiol. 2020, 86, e01769-20.
- Mafa, M.S.; Malgas, S.; Rashamuse, K.; Pletschke, B.I. Delineating functional properties of a cello-oligosaccharide and β-glucan specific cellobiohydrolase (GH5_38): Its synergism with Cel6A and Cel7A for β-(1,3)-(1,4)-glucan degradation. Carbohydr. Res. 2020, 495, 108081.
- Carli, S.; Carneiro, A.L.C.B.; Ward, R.J.; Meleiro, L.P. Immobilization of a β-glucosidase and an endoglucanase in ferromagnetic nanoparticles: A study of synergistic effects. Protein Expr. Purif. 2019, 160, 28–35.
- Uchiyama, T.; Uchihashi, T.; Nakamura, A.; Watanabe, H.; Kaneko, S.; Samejima, M.; Igarashi, K. Convergent evolution of processivity in bacterial and fungal cellulases. Proc. Natl. Acad. Sci. USA 2020, 117, 19896–19903.
- Forsberg, Z.; Vaaje-Kolstad, G.; Westereng, B.; Bunæs, A.C.; Stenstrøm, Y.; MacKenzie, A.; Sørlie, M.; Horn, S.J.; Eijsink, V.G.H. Cleavage of cellulose by a CBM33 protein. Protein Sci. 2011, 20, 1479–1483.
- Bunterngsook, B.; Mhuantong, W.; Champreda, V.; Thamchaipenet, A.; Eurwilaichitr, L. Identification of novel bacterial expansins and their synergistic actions on cellulose degradation. Bioresour. Technol. 2014, 159, 64–71.
- Isaksen, T.; Westereng, B.; Aachmann, F.L.; Agger, J.W.; Kracher, D.; Kittl, R.; Ludwig, R.; Haltrich, D.; Eijsink, V.G.H.; Horn, S.J. A c4-oxidizing lytic polysaccharide monooxygenase cleaving both cellulose and cello-oligosaccharides. J. Biol. Chem. 2014, 289, 2632–2642.
- Kadowaki, M.A.S.; Magri, S.; de Godoy, M.O.; Monclaro, A.V.; Zarattini, M.; Cannella, D. A fast and easy strategy for lytic polysaccharide monooxygenase-cleavable His6-Tag cloning, expression, and purification. Enzym. Microb. Technol. 2021, 143, 109704.
- Saloheimo, M.; Paloheimo, M.; Hakola, S.; Pere, J.; Swanson, B.; Nyyssonen, E.; Bhatia, A.; Ward, M.; Penttila, M. Swollenin, a Trichoderma reesei protein with sequence similarity to the plant expansins, exhibits disruption activity on cellulosic materials. Eur. J. Biochem. 2002, 269, 4202–4211.
- Lee, H.J.; Lee, S.; Ko, H.; Kim, H.K.; Choi, I. An expansin-like protein from Hahella chejuensis binds cellulose and enhances cellulase activity. Mol. Cells 2010, 29, 379–385.
- Junior, A.T.; Dolce, L.G.; Neto, M.O.; Polikarpov, I. Xanthomonas campestris expansin-like X domain is a structurally disordered beta-sheet macromolecule capable of synergistically enhancing enzymatic efficiency of cellulose hydrolysis. Biotechnol. Lett. 2015, 37, 2419–2426.
- Boisset, C.; Fraschini, C.; Schulein, M.; Henrissat, B.; Chanzy, H. Imaging the enzymatic digestion of bacterial cellulose ribbons reveals the endo character of the cellobiohydrolase Cel6A from Humicola insolens and its mode of synergy with cellobiohydrolase Cel7A. Appl. Environ. Microbiol. 2000, 66, 1444–1452.
- Zhang, X.-Z.; Sathitsuksanoh, N.; Zhang, Y.-H.P. Glycoside hydrolase family 9 processive endoglucanase from Clostridium phytofermentans: Heterologous expression, characterization, and synergy with family 48 cellobiohydrolase. Bioresour. Technol. 2010, 101, 5534–5538.
- Tang, Z.; Li, P.; Sun, R.; Liu, M.; Jin, W.; Gou, L.; Chen, H.; Wu, Q.; Bu, T.; Li, C. Optimum mixture and synergy analysis of three main cellulases. Int. J. Agric. Biol. 2018, 20, 1814–9596.
- Zhou, S.; Ingram, L.O. Synergistic hydrolysis of carboxymethyl cellulose and acid-swollen cellulose by two endoglucanases (CelZ and CelY) from Erwinia chrysanthemi. J. Bacterial. 2000, 182, 5676–5682.
- Tuveng, T.R.; Jensen, M.S.; Fredriksen, L.; Vaaje-Kolstad, G.; Eijsink, V.G.H.; Forsberg, Z. A thermostable bacterial lytic polysaccharide monooxygenase with high operational stability in a wide temperature range. Biotechnol. Biofuels 2020, 13, 194.
- Yang, M.; Zhang, K.; Zhang, P.; Zhou, X.; Ma, X.; Li, F. Synergistic cellulose hydrolysis dominated by a multi-modular processive endo glucanase from Clostridium cellulosi. Front. Microbiol. 2016, 7, 932.
- Zhou, H.; Li, T.; Yua, Z.; Jua, J.; Zhanga, H.; Tan, H.; Li, K.; Yin, H. A lytic polysaccharide monooxygenase from Myceliophthora thermophila and its synergism with cellobiohydrolases in cellulose hydrolysis. Int. J. Biol. Macromol. 2019, 139, 570–576.
- Karnaouri, A.; Muraleedharan, M.N.; Dimarogona, M.; Topakas, E.; Rova, U.; Sandgren, M.; Christakopoulos, P. Recombinant expression of thermostable processive MtEG5 endoglucanase and its synergism with MtLPMO from Myceliophthora thermophila during the hydrolysis of lignocellulosic substrates. Biotechnol. Biofuels 2017, 10, 126.
- Zverlov, V.; Mah, S.; Riedel, K.; Bronnenmeier, K. Properties and gene structure of a bifunctional cellulolytic enzyme (CelA) from the extreme thermophile ‘Anaerocellum thermophilum’ with separate glycosyl hydrolase family 9 and 48 catalytic domains. Microbiology 1998, 144, 457–465.
- Malgas, S.; Thoresen, M.; van Dyk, J.S.; Pletschke, B.I. Time dependence of enzyme synergism during the degradation of model and natural lignocellulosic substrates. Enzym. Microb. Technol. 2017, 103, 1–11.
- Tokin, R.; Ipsen, J.O.; Westh, P.; Johansen, K.S. The synergy between LPMOs and cellulases in enzymatic saccharification of cellulose is both enzyme- and substrate-dependent. Biotechnol. Lett. 2020, 42, 1975–1984.
- Oh, C.H.; Park, C.S.; Lee, Y.G.; Song, Y.; Bae, H.-J. Characterization of acidic endoglucanase Cel12A from Gloeophyllum trabeum and its synergistic effects on hydrogen peroxide–acetic acid (HPAC)-pretreated lignocellulose. J. Wood Sci. 2019, 65, 24.
- Lutzen, N.W.; Nielsen, M.K.; Oxenboell, K.M.; Schtlein, M.; Stentebjerg-olesen, B. Cellulases and their application in the conversion of lignocellulose to fermentable sugars. Phil. Trans. R. Soc. Lond. B 1983, 300, 283–291.
- Liu, Y.; Nemmaru, B.; Chundawat, P.S. Thermobifida fusca cellulases exhibit increased endo−exo synergistic activity, but lower exocellulase activity, on cellulose-III. ACS Sustain. Chem. Eng. 2020, 8, 5028–5039.
- Liu, Y.; Nemmaru, B.; Chundawat, P.S. Thermobifida fusca cellulases exhibit increased endo−exo synergistic activity, but lower exocellulase activity, on cellulose-III. ACS Sustain. Chem. Eng. 2020, 8, 5028–5039.
- Pellegrini, V.O.; Bernardes, A.; Rezende, C.A.; Polikarpov, I. Cellulose fiber size defines efficiency of enzymatic hydrolysis and impacts degree of synergy between endo and exoglucanases. Cellulose 2018, 25, 1865–1881.
- Woodward, J.; Lima, M.; Lee, N.E. The role of cellulase concentration in determining the degree of synergism in the hydrolysis of microcrystalline cellulose. Biochem. J. 1988, 255, 895–899.
- Shi, J.; Wu, D.; Zhang, L.; Simmons, B.A.; Singh, S.; Yang, B.; Wyman, C.E. Dynamic changes of substrate reactivity and enzyme adsorption on partially hydrolyzed cellulose. Biotechnol. Bioeng. 2017, 114, 503–515.
- Eriksson, T.; Karlsson, J.; Tjerneld, F. A model explaining declining rate in hydrolysis of lignocellulose substrates with cellobiohydrolase I (Cel7A) and endoglucanase I (Cel7B) of Trichoderma rees. Appl. Biochem. Biotechnol. 2002, 101, 42–58.
- Filiatrault-Chastel, C.; Navarro, D.; Haon, M.; Grisel, S.; Herpoël-Gimbert, I.; Chevret, D.; Fanuel, M.; Henrissat, B.; Heiss-Blanquet, S.; Margeot, A.; et al. AA16, a new lytic polysaccharide monooxygenase family identified in fungal secretomes. Biotechnol. Biofuels 2019, 12, 55.
- Tuveng, T.R.; Jensen, M.S.; Fredriksen, L.; Vaaje-Kolstad, G.; Eijsink, V.G.H.; Forsberg, Z. A thermostable bacterial lytic polysaccharide monooxygenase with high operational stability in a wide temperature range. Biotechnol. Biofuels 2020, 13, 194.
- Tokin, R.; Ipsen, J.O.; Westh, P.; Johansen, K.S. The synergy between LPMOs and cellulases in enzymatic saccharification of cellulose is both enzyme- and substrate-dependent. Biotechnol. Lett. 2020, 42, 1975–1984.
- Zhou, H.; Li, T.; Yua, Z.; Jua, J.; Zhanga, H.; Tan, H.; Li, K.; Yin, H. A lytic polysaccharide monooxygenase from Myceliophthora thermophila and its synergism with cellobiohydrolases in cellulose hydrolysis. Int. J. Biol. Macromol. 2019, 139, 570–576.
- Keller, N.B.; Badino, F.S.; Røjel, N.; Sørensen, T.H.; Kari, J.; McBrayer, B.; Borch, K.; Blossom, B.M.; Westh, P. A comparative biochemical investigation of the impeding effect of C1-oxidizing LPMOs on cellobiohydrolases. J. Biol. Chem. 2021, 296, 100504.
- Karnaouri, A.; Muraleedharan, M.N.; Dimarogona, M.; Topakas, E.; Rova, U.; Sandgren, M.; Christakopoulos, P. Recombinant expression of thermostable processive MtEG5 endoglucanase and its synergism with MtLPMO from Myceliophthora thermophila during the hydrolysis of lignocellulosic substrates. Biotechnol. Biofuels 2017, 10, 126.
- Santos, C.A.; Ferreira-Filho, J.A.; O’Donovan, A.; Gupta, V.K.; Maria, G.; Tuohy, M.G.; Souza, A.P. Production of a recombinant swollenin from Trichoderma harzianum in Escherichia coli and its potential synergistic role in biomass degradation. Microb. Cell Fact. 2017, 16, 83.
- Malgas, S.; Thoresen, M.; van Dyk, J.S.; Pletschke, B.I. Time dependence of enzyme synergism during the degradation of model and natural lignocellulosic substrates. Enzym. Microb. Technol. 2017, 103, 1–11.
- Boisset, C.; Petrequin, C.; Chanzy, H.; Henrissat, B.; Schulein, M. Optimized mixtures of recombinant Humicola insolens cellulases for the biodegradation of crystalline cellulose. Biotechnol. Bioeng. 2001, 72, 3.
- Zverlov, V.; Mah, S.; Riedel, K.; Bronnenmeier, K. Properties and gene structure of a bifunctional cellulolytic enzyme (CelA) from the extreme thermophile ‘Anaerocellum thermophilum’ with separate glycosyl hydrolase family 9 and 48 catalytic domains. Microbiology 1998, 144, 457–465.
- Lutzen, N.W.; Nielsen, M.K.; Oxenboell, K.M.; Schtlein, M.; Stentebjerg-olesen, B. Cellulases and their application in the conversion of lignocellulose to fermentable sugars. Phil. Trans. R. Soc. Lond. B 1983, 300, 283–291.
- Oh, C.H.; Park, C.S.; Lee, Y.G.; Song, Y.; Bae, H.-J. Characterization of acidic endoglucanase Cel12A from Gloeophyllum trabeum and its synergistic effects on hydrogen peroxide–acetic acid (HPAC)-pretreated lignocellulose. J. Wood Sci. 2019, 65, 24.
