2. General Features and Roles of Long Non-Coding RNAs
LncRNAs are non-protein-coding RNA species that are around 200 base pair or more in size. The recent annotations estimate their numbers to be around 31,678 in humans but this number may exceed [
8,
9,
10,
11]. However, how many of these lncRNAs are functional is still not known. Compared to protein coding genes, lncRNAs display highly lineage-restricted expression and show poor conservation at the sequence level. On the contrary, lncRNA promoters show a high level of conservation. However, emerging evidence indicates that the primary conservation in lncRNAs is more intricate and should not be visualized only from the sequence perspective. LncRNAs can be conserved at multiple levels such as secondary or higher-order tertiary structure, binding motifs for interacting proteins, functional features, and genomic location [
12,
13]. In addition to structural complexity, lncRNAs adopt diverse molecular mechanisms for executing their regulatory functions. They can act as scaffolds for targeting chromatin modifying enzymes, transcriptional factors, and regulators of splicing, and as sponges or decoys for microRNAs and transcription factors, etc. Details of molecular mechanisms of gene regulation by lncRNAs have been discussed in greater detail in a number of reviews [
1,
2,
14].
3. Understanding the Contribution of lncRNA in Cancer Development
Tumours, in general, are comprised of heterogeneous cells with varying cell states. A combination of alterations in genetic, signalling, metabolic, and microenvironments converge to dictate the fate of tumour progression. According to the protein-centric view of cancer development, mutations/alterations in key genes such as
EGFR,
ALK,
BRAF,
KRAS, TP53, and
MYC mainly drive the oncogenic events and thus influence the possible clinical outcomes [
15,
16,
17,
18,
19]. However, this information is insufficient as the cancer treatments based on protein-coding genes provide a limited cure, especially for relapsed cancer patients. As the non-coding genome has been gaining traction lately among researchers due to its surprising functions, it is pertinent to examine its contribution to cancer development. In fact, it is vital to analyse if lncRNAs can also work as master regulators of tumorigenesis or if they are just followers of protein-coding master regulators. In the current section, we will examine this statement in light of well-known lncRNAs reported in more than one cancer and associated with key aspects of cancer development (
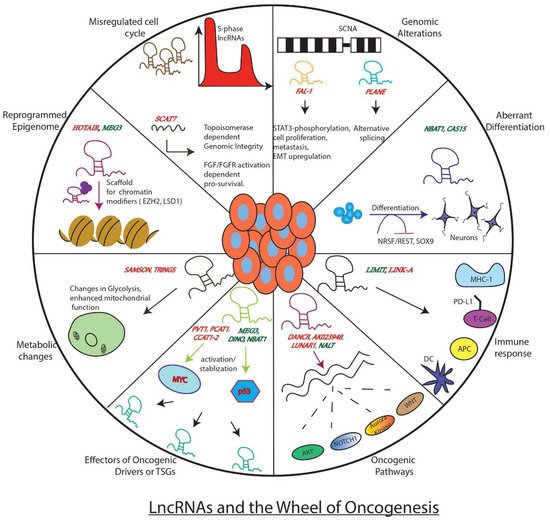
Figure 1).
Figure 1. Well known examples of LncRNAs and how they influence key genes, processes, or pathways involved in tumor development. The figure depicts the regulatory role of lncRNAs as either promoters of the wheel of oncogenesis (red) or as brakes (green) inhibiting its movement.
3.1. LncRNA in Oncogenic Pathways
LncRNAs can act as promoters of key oncogenic events associated with cancer development. These events include cell growth, invasion, metastasis, suppression of cell death pathway, alteration in DNA damage response, etc.
HOTAIR is one such lncRNA reported in oncogenic roles across multiple cancer types. Moreover, it can act as an independent predictor of clinical outcome and thus is suggested as an ideal biomarker for poor prognosis. It is known to regulate its downstream effectors via targeting well-known repressive chromatin modifiers, Polycomb Repressive Complex 2 (PRC2), and LSD1 complex (H3K4 demethylase) to its target promoters [
20]. This
HOTAIR-induced reprogrammed epigenome, in turn, promotes the gene expression pattern required for breast cancer metastasis. Additionally, it can function as a competitive endogenous RNA (ceRNA) and regulate the critical micro-RNA dependent TGF-β, PI3K-AKT, Wnt/β, and VEGF oncogenic-signalling cascades [
21,
22].
Another oncogenic lncRNA is the leukemia-induced non-coding activating RNA 1 (
LUNAR1), initially identified as a critical downstream target of oncogenic Notch1 in T cell acute lymphoblastic leukaemia (T-ALL) [
23]. It executes its oncogenic regulatory function by controlling the activation of insulin-like growth receptor 1 (IGF1R), a Notch1 target that mediates growth/survival signals in T-ALL.
LUNAR1 functions as an enhancer-like lncRNA whereby it recruits mediator complex to the intronic
IGF1R enhancer and thus fully activates
IGF1R promoter via DNA looping. As oncogenic Notch1 and over-activation of IGF1 are known to promote cancer development,
LUNAR1 may be active in other cancers with Notch1 alterations. Indeed, its elevated expression was found to be associated with colorectal cancer aggressiveness, and thus this lncRNA may work as a promising prognostic marker as well as a therapeutic target for tumours driven by altered Notch1 signalling. Another lncRNA,
NALT, is also reported as a potential activator of NOTCH1 [
24]. Other pro-oncogenic lncRNAs, such as
DANCR and
AK023948 are known to function as positive regulators of AKT pathway [
25,
26].
Recent studies have turned to cancer cell hallmark-based screens to identify tumor driving lncRNAs. For example, deregulated cell cycle, one of the well-studied cancer cell hallmarks, has been used to identify lncRNA-based oncogenic drivers. Using an S-phase based nascent capture assay, a recent investigation has identified 570 S-phase enriched lncRNAs that show significant differential expression in at least one tumor type across pan-cancer TCGA data sets. Mechanistic investigations on one of the top S-phase enriched cancer associated lncRNAs
SCAT7 (
ELF3-AS1) revealed that it promotes tumorigenesis through activating fibroblast growth factor (FGF/FGFR)-dependent PI3K/AKT and MAPK pro-survival pathways via interacting with hnRNPK/YBX1 complex [
4]. Besides, it maintains genome integrity through regulating Topoisomerase 1 turnover [
27]. Thus, S-phase enriched cancer associated lncRNAs will serve as an interesting resource for identifying potential new targets for cancer treatment.
Another intelligent approach that has yielded identifications of lncRNAs in oncogenic roles has been the genomic and transcriptomic analysis of somatic copy number alterations (SCNAs), frequently observed across tumour types. SCNA analysis revealed two interesting lncRNAs,
FAL-1 and
PLANE, that, like their protein-coding counterparts at amplified regions (such as
MYCN gene amplification in neuroblastoma tumors), have been functionally characterized for their tumour-promoting roles [
28,
29].
FAL-1 maps to the frequently amplified region on chromosome 1, and its dysregulation is observed across multiple tumour types. Moreover, its association with poor prognosis makes it an attractive candidate for disease prognosis and targeted therapy.
FAL-1 exerts its oncogenic functions, like promoting cell proliferation and metastasis via diverse regulatory mechanisms such as stabilising epigenetic repressor BMI 1, enhancer-like functions, phosphorylation of STAT3, and upregulation of EMT proteins. Similar to
FAL-1,
PLANE maps to the chromosome 3q region and promotes oncogenic pathways by regulating an alternative splicing programme [
29].
Apart from their more direct function, lncRNAs can also work either as modulators or downstream effectors of oncogenic drivers. This is best seen in the case of the known transcription factor and oncogenic driver,
MYC. LncRNAs such as
PVT1,
PCAT1,
CCAT1, and
CCAT2 affect the expression of
MYC, where
PVT1 post-transcriptionally stabilizes MYC,
PCAT1 functions as a ceRNA preventing the downregulation of MYC by miR-34a [
30,
31].
CCAT1 and
CCAT2 are the two enhancer-driven lncRNAs located upstream of the
MYC gene that influences its expression via cis-regulating mechanisms [
32,
33]. In addition to these upstream regulators of
MYC, 19 lncRNAs as downstream effectors of
MYC have been reported [
34].
All these examples clearly indicate how oncogenic lncRNAs are instrumental in the process of carcinogenesis. By functioning as modulators of epigenetic machinery, signalling pathways, long-range chromatin interactions, oncogenes, and micro-RNA functions, lncRNAs seem to play a significant role in the oncogenic process. Due to their elevated expression and association with unfavourable clinical outcomes, oncogenic lncRNAs are ideal candidates for prognostic markers and make attractive targets for therapeutic interventions. Although some of the lncRNAs, such as HOTAIR, have been extensively investigated as oncogenic drivers in multiple cancer types, we are still far from understanding the major common oncogenic pathways by which they induce neoplastic transformation. Thus, more work is needed on these lines to develop lncRNAs as more reliable targets for cancer therapy.
3.2. LncRNAs in Tumor Suppressor Pathways
In addition to their role in promoting tumour development, lncRNAs are also known to be involved in tumour suppressive roles. We will discuss some well-established lncRNAs which are exclusively known to function as suppressors of tumour development across many cancers. Further, we will extrapolate these findings in speculating whether lncRNA work as bonafide tumour suppressors or just as assistants to protein-coding TSGs (tumour suppressor genes).
Well-studied examples of tumour suppressor lncRNAs are known in Neuroblastoma (NB), an embryonal childhood cancer of peripheral nervous system. Initial genome-wide association studies (GWAS) in these tumours uncovered a risk locus 6p22.3 with clusters of risk-associated single nucleotide polymorphisms (SNPs). What is more fascinating is that these SNPs are present at a genomic location with no protein-coding genes. These genomic hot spots consist of sense and antisense pairs of lncRNAs
CAS15 and
NBAT1, whose reduced levels are observed in high-risk NBs and are associated with poor prognosis [
35,
36]. The lower expression of sense
CASC-15 and antisense
NBAT1 in high-risk NBs is caused due to epigenetic suppression of their promoter.
NBAT1 exerts its tumour suppressive activity by epigenetic silencing of genes such as
SOX9,
VCAN, and
OSMR via its association with chromatin repressor, EZH2 [
37,
38]. It also promotes neuronal differentiation by modulating neuronal-specific transcription factor NRSF/REST.
NBAT1’s sense counterpart
CASC15 has several isoforms and among which
CASC15-003 and
CASC15-004 have been shown to display tumor suppressor functions. Of note, the isoform
CASC15-003 shares common biological pathways with
NBAT1 in promoting tumor suppression and inducing neuronal differentiation. For example, both the sense (
CASC15-003) and antisense (
NBAT1) lncRNAs regulate the expression of SOX9, a well-characterized neural crest-specific transcription factor, through controlling CHD7 stability by modulating the nucleolar-specific localization of deubiquitinating enzyme USP36 by interfering with USP36 and NPM1 interaction. Consistent with these observations,
NBAT1 and
CASC15-003 rescue each other’s functions in retinoic acid induced neuronal differentiation, thus these lncRNAs are functionally complement each in promoting their functions. These data also indicate that
NBAT1 controls SOX9 expression at the transcriptional and post-transcriptional level, whereas
CASC15 regulates SOX9 expression at the transcriptional level. Thus, SOX9 appears to be one of the key genes in neuroblastoma progression, whose oncogenic functions are controlled by
NBAT1 and
CASC15-003. Their optimal levels are essential to ensure normal neuronal differentiation and suppressing cancer progression and development. The broader clinical significance of these lncRNAs is reflected by their identification as prognostic markers in other cancers. For instance,
NBAT1 is a potential tumor suppressor, and its higher expression is associated with good clinical outcomes in cancers such as NSCLC, oesophageal cancer, HCC, renal cell carcinoma, and breast cancer [
39,
40,
41,
42]. Thus,
NBAT1 is a bona fide TSG.
NORAD (non-coding RNA activated by DNA damage) is another candidate lncRNA with tumour-suppressive properties, initially identified in DNA damage screen in colon cancer cell lines. It is a conserved, cytoplasmic, localised, abundant RNA with multiple repeat sequences believed to be assembly platforms of Pumilio ribonuclear protein complexes (PUM1 and PUM2). The lncRNA
NORAD prevents aberrant mitosis by inhibiting Pumilio (PUM) proteins [
43]. Its TSG functions mainly occur as a decoy for PUM1 and PUM2 by nucleating the formation of phase-separated Pumilio condensates, termed NP bodies [
44]. Disruption of NORAD-mediated PUM phase separation leads to hyperactivation of Pumulio proteins and genome instability. Similarly, it is also reported to sequester pro-metastatic regulator, S100-P, thus suppressing metastasis in breast and lung cancers [
45].
GAS5 (Growth Arrest-Specific Transcript 5) is also an interesting tumour suppressor lncRNA whose lowered expression is associated with poorer clinical outcomes in a variety of human malignancies [
46]. It has been functionally implicated in wide cancer-related processes such as cell proliferation, invasion, metastasis, epithelial to mesenchymal transition (EMT), drug resistance, etc. [
47]. This plethora of functions are carried out using diverse mechanisms such as association with chromatin-modifying enzymes, acting as a sponge for mi-RNAs and formation of DNA-RNA triplex structures.
Like
GAS5,
MEG3, an imprinted lncRNA with maternal-specific expression, has been shown to act as a tumor suppressor in several cancers by being in strong functional connection with epigenetic machinery. Functional investigations have shown that this lncRNA suppresses tumors through downregulating oncogenic pathways such as MYC and TGF-β and activating p53 and RB. Published data indicates that
MEG-3-dependent suppression of oncogenic pathways involves large-scale epigenetic programming.
MEG3 has been shown to have distinct interactions with the PRC2 complex and its cofactor, JARID2. Such interactions have been implicated in the regulation of expression of transforming growth factor-β (TGF-β) pathway genes. Further mechanistic investigations revealed that
MEG-3 suppresses TGF-β pathway genes through forming triplexes at purine-rich GA stretches to recruit chromatin modifiers [
48].
LncRNA molecules also exert their tumour-suppressive functions by either inhibiting cancer promoting oncogenic pathways or fine-tuning the response of protein coding TSGs.
NKILA (NF-κB interacting LncRNA) is one such tumour-suppressive RNA that works as a feedback regulator of NF-κB activity [
49]. Its decreased expression is associated with poor outcomes in patients.
NKILA acts as a scaffold to recruit NF-κB/IκB complex and prevents phosphorylation of IκB by IKK. This RNA is upregulated by NF-κB and forms a negative feedback loop of NF-κB. Additionally, a mouse lncRNA,
Lethe, is induced by NF-κB and functions as a negative feedback regulator of NF-κB [
50]. However, the significance of
Lethe in cancer remains unknown. LncRNAs’ function as fine-tuners of TSG activity is best exemplified in the case of the well-known TSG,
TP53 [
51]. Several lncRNAs are reported as components of a p53-mediated network where they work either as its modulators or downstream effectors. Decreased expression of maternally expressed genes 3 (
MEG3) lncRNA is seen in many cancers, and it is known to activate p53 and thus modulate its downstream target activity [
52].
MEG-3 functional connection with p53 has been well established by several studies. A recent work has characterized conserved pseudoknots in
MEG3 that are responsible for the p53 stimulation, revealing the structural basis for the p53 stimulation [
53]. Although
MEG-3 has been shown to directly interact with p53 protein, several studies have implicated the indirect role of
MEG-3 in p53 pathway stimulation [
54]. Thus, functional dissection of this well-connected tumor suppressor pathway will help in uncovering the molecular basis of MEG-3-p53-dependent tumor suppression. Another lncRNA induced by DNA damage is conserved lncRNA,
DINO (damage induced non-coding), present upstream of
CDKN1A [
55]. In response to DNA damage, it binds and stabilizes p53 and thus activates the p53-mediated gene network. Its functional relevance is elucidated in the transgenic mice (knockout or inactivated promoter) of
DINO showing dampened p53-induced DNA damage response. Along similar lines,
NBAT1 is a p53-responsive lncRNA, that in turn regulates p53 subcellular levels through controlling p53 nuclear export machinery CRM1. Higher
NBAT1 expression correlates with increased p53 signaling, and depletion of
NBAT1 altered CRM1 function, contributing to the loss of p53-dependent nuclear gene expression [
56]. Based on NBAT1-CRM1-p53 functional connection, Nutlin-3a and Selinexor treatment has been proposed for the treatment of Neuroblastoma patients. On the other hand, lncRNAs such as
lincRNA-p21, Lincprint,
Tug1, and
PANDA form the downstream components of the p53 network where
lincRNA-p21 and
Lincprint exert their p53 tumor suppressive function via association with hnRNPK and PRC2, respectively.
PANDA, p53-inducible lncRNA, sequesters and regulate NFYA (nuclear transcription factor Y, alpha) levels to control the expression of pro-survival and pro-apoptotic genes [
51].
3.3. Context-Specific Roles of LncRNAs as Oncogenes or TSGs
It is evident from the differential expression analyses of lncRNAs in pan-cancer TCGA data analyses that several lncRNAs display both oncogenic or tumor suppressive functions in a cancer-specific fashion. For example,
H19 has been shown to possess oncogenic properties in multiple cancers such as colorectal cancer, breast cancer and hepatocellular carcinoma, whereas in human rhabdoid cancers,
H19 displays tumour-suppressive roles [
57,
58].
Similar observations have been reported for another lncRNA,
NEAT1, which is involved in paraspeckle formation. Although it is mostly reported to possess oncogenic roles in multiple cancers such as glioma, ovarian cancer, melanoma and prostate cancer, its loss has been shown to promote tumorigenesis in a mouse model of pancreatic ductal adenocarcinoma [
59,
60,
61]. Another lncRNA that displays context-specific function as an oncogene or tumor suppressor is
CASC15, which has been shown to act as an oncogene in skin cancers whereas as a tumor suppressor in the context of Neuroblastoma [
36,
62].
TINCR (terminal differentiation-induced non-coding RNA) is another example of lncRNAs with dual functions in the progression of cancer. While it adopts a tumor suppressive role in glioma, prostate cancer and retinoblastoma; it works as an oncogene in gastric, bladder and breast cancer [
21].
These examples indicate towards dynamic nature of lncRNA molecules which can adopt contrasting roles in a cell of the origin-specific manner in tumour development. Additionally, the tumor microenvironment could also influence the functional role of a given lncRNA in a genetic context. Do such lncRNAs have special structural or regulatory features that provide them the adaptability in a given tumor context remains unknown? Finally, treatment approaches that could alter the expression of such lncRNAs should be used with caution.
3.4. LncRNAs Influence Tumorigenesis through Metabolic Pathways
Metabolic reprogramming is one of the key hallmarks associated with cancers. In addition to protein coding genes, lncRNAs are also reported to participate in regulation of metabolic pathways. For example, lncRNA
SAMSON promotes melanoma survival by enhancing mitochondrial function [
63]. It regulates the maturation of mitochondrial 16S rRNA via interaction with p32. Overexpression of
SAMSON creates a unique metabolic dependency in melanoma cells; targeting the lncRNA leads to reduced growth/viability of invasive melanoma cells. Another lncRNA, Tp53-regulated inhibitor of necrosis (
TRINGS) gets elevated after glucose deprivation in cancer cells.
TRINGS promotes tumour growth (in vitro and in vivo) via binding to STRAP and inhibits the STRAP-GSK3β-NF-κB necrotic pathway [
64]. Other lncRNAs such as
lincRNA-p21 and FoxO-induced long non-coding RNA 1 (
FILNC1) are also involved in regulating glycolysis [
65,
66]. HIF-1α induced
lincRNA-p21 disrupts VHL-HIF-1α interaction by binding to both the molecules and leads to a collection of HIF-1α, and thus hypoxia-enhanced glycolysis. On the other hand,
FILNC1 is a translation suppressor of c-Myc RNA, and thus, its downregulation enhances the Warburg effect through c-Myc upregulation. Contrary to
FILNC1, using siRNA screen, Sang et al. have identified a lncRNA for calcium-dependent kinase activity (CamK-A) that promotes the Warburg effect. This lncRNA is highly expressed in various types of cancer and is involved in remodelling the tumour microenvironment via activation of calcium trigged signalling [
67]. LncRNAs can also act as signalling molecules that transmit signals between immune and tumour cells to promote the Warburg effect. For instance, Chen et al. have reported that tumor-associated macrophages (TAM) enhances aerobic glycolysis via synthesis and secretion of an extracellular vesicle packaged HIF-1α-stabilizing long non-coding RNA (
HISLA) [
68]. Taken together, lncRNAs play crucial roles in regulating metabolism and consequently impact the associated process of the tumor microenvironment.