CRISPR activation (CRISPRa) is one type of CRISPR tool that use modified versions of dCas9, a mutation of Cas9 without endonuclease activity, with added transcriptional activators on dCas9 or the guide RNAs (gRNAs). Like a standard CRISPR-Cas9 system, dCas9 activation systems rely on similar components such as Cas9 variants for modulation or modification of genes, gRNAs to guide Cas9 to intended targets, and vectors for introduction into cells. However, while a standard CRISPR-Cas9 system relies on creating breaks in DNA through the endonuclease activity of Cas9 and then manipulating DNA Repair mechanisms for gene editing, dCas9 activation systems are modified and employ transcriptional activators to increase expression of genes of interest. Such systems are usable for many purposes including but not limited to, genetic screens and overexpression of proteins of interest.
- endonuclease activity
- endonuclease
- dcas9
1. Components
1.1. dCas9
Cas9 Endonuclease Dead, also known as dead Cas9 or dCas9, is a mutant form of Cas9 whose endonuclease activity is removed through point mutations in its endonuclease domains. Similar to its unmutated form, dCas9 is used in CRISPR systems along with gRNAs to target specific genes or nucleotides complementary to the gRNA with PAM sequences that allow Cas9 to bind. Cas9 ordinarily has 2 endonuclease domains called the RuvC and HNH domains. The point mutations D10A and H840A change 2 important residues for endonuclease activity that ultimately results in its deactivation. Although dCas9 lacks endonuclease activity, it is still capable of binding to its guide RNA and the DNA strand that is being targeted because such binding is managed by other domains. This alone is often enough to attenuate if not outright block transcription of the targeted gene if the gRNA positions dCas9 in a way that prevents transcriptional factors and RNA polymerase from accessing the DNA. However, this ability to bind DNA can also be exploited for activation since dCas9 has modifiable regions, typically the N and C terminus of the protein, that can be used to attach transcriptional activators.[1]
1.2. Guide RNA
A small guide RNA (sgRNA), or gRNA is an RNA with around 20 nucleotides used to direct Cas9 or dCas9 to their targets. gRNAs contain two major regions of importance for CRISPR systems: the scaffold and spacer regions. The spacer region has nucleotides that are complementary to those found on the target genes, often in the promoter region. The scaffold region is responsible for formation of a complex with (d)Cas9. Together, they bind (d)Cas9 and direct it to the gene(s) of interest. Since the spacer region of a gRNA can be modified for any potential sequence, they give CRISPR systems much more flexibility as any genes and nucleotides with a sequence complementary to the spacer region can become possible targets.[1]
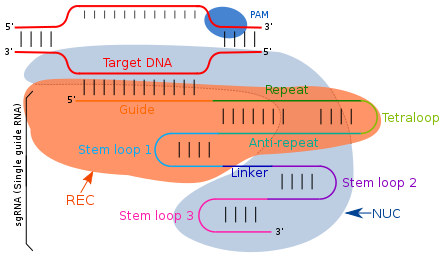
1.3. Transcriptional Activators
Transcriptional Activators are protein domains or whole proteins linked to dCas9 or sgRNAs that assist in the recruitment of important co-factors as well as RNA Polymerase for transcription of the gene(s) targeted by the system. In order for a protein to be made from the gene that encodes it, RNA polymerase must make RNA from the DNA template of the gene during a process called transcription. Transcriptional activators have a DNA binding domain and a domain for activation of transcription. The activation domain can recruit general transcription factors or RNA polymerase to the gene sequence. Activation domains can also function by facilitating transcription by stalled RNA polymerases, and in eukaryotes can act to move nucleosomes on the DNA or modify histones to increase gene expression.[2] These activators can be introduced into the system through attachment to dCas9 or to the sgRNA. Some researchers have noted that the extent of transcriptional upregulation can be modulated by using multiple sites for activator attachment in one experiment and by using different variations and combinations of activators at once in a given experiment or sample.[3][4][5]
1.4. Expression System
An expression system is required for the introduction of the gRNAs and (d)Cas9 proteins into the cells of interest. Typically employed options include but are not limited to plasmids and viral vectors such as adeno-associated virus (AAV) vector or lentivirus vector.
2. Specific Activation Systems
2.1. VP64-p65-Rta
The VP64-p65-Rta, or VPR, dCas9 activator was created by modifying an existing dCas9 activator, in which a Vp64 transcriptional activator is joined to the C terminus of dCas9. In the dCas9-VPR protein, the transcription factors p65 and Rta are added to the C terminus of dCas9-Vp64. Therefore, all three transcription factors are targeted to the same gene. The use of three transcription factors, as opposed to solely Vp64, results in increased expression of targeted genes. When different genes were targeted by dCas9, they all showed significantly greater expression with dCas9-VPR than with dCas9-VP64. It has also been demonstrated that dCas9-VPR can be used to increase expression of multiple genes within the same cell by putting multiple sgRNAs into the same cell.[6] dCas9-VPR has been used to activate the neurogenin 2 (link) and neurogenic differentiation 1 (link) genes, resulting in differentiation of induced pluripotent stem cells into induced neurons.[6] A study comparing dCas9 activators found that the VPR, SAM, and Suntag activators worked best with dCas9 to increase gene expression in a variety of fruit fly, mouse, and human cell types.[7]
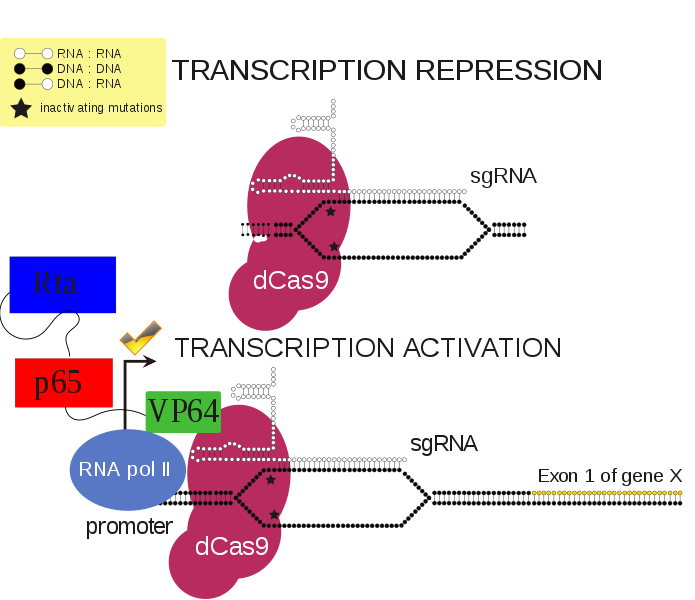
2.2. Synergistic Activation Mediator
To overcome the limitation of the dCas9-VP64 gene activation system, the dCas9-SAM system was developed to incorporate multiple transcriptional factors. Utilizing MS2, p65, and HSF1 proteins, dCas9-SAM system recruits various transcriptional factors working synergistically to activate the gene of interest.
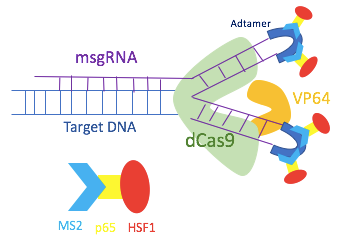
In order to assemble different transcriptional activators, the dCas9-SAM system uses a modified single guide RNA (sgRNA) that has binding sites for the MS2 protein. Hairpin aptamers are attached to the tetra loop and the stem loop 2 of the sgRNA to become binding sites for dimerized MS2 bacteriophage coat proteins. As the hairpins are exposed outside of the dCas9-sgRNA complex, other transcriptional factors can bind to the MS2 protein without disrupting the dCas9-sgRNA complex. Thus, the MS2 protein is engineered to include p65 and HSF1 proteins. The MS2-p65-HSF1 fusion protein interacts with the dCas9-VP64 to recruit more transcriptional factors onto the promoter of the target genes.
Employing the dCas-SAM system, Zhang et al. (2015) successfully reactivated the latent HIV gene to over-express viral proteins from the HIV host cells.[8] They were able to over-express viral proteins substantially to trigger apoptosis of HIV-1 latent cells due to the toxicity of viral proteins. In another dCas-SAM system experiment, Konermann et al. (2015) found genes in melanoma cells that give resistance to a BRAF inhibitor through activating candidate genes via dCas system.[5] Thus, the dCas9-SAM system can further be employed to activate latent genes, develop gene therapies, and discover new genes.
2.3. SunTag
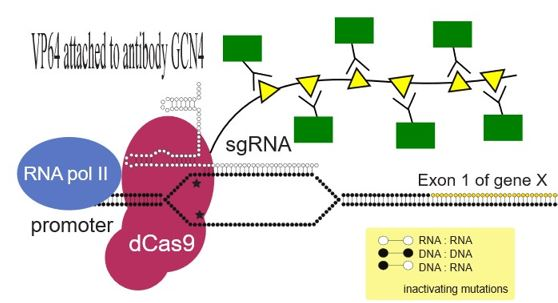
The SunTag activator system uses the dCas9 protein, which is modified to be linked with the SunTag. The SunTag is a repeating polypeptide array that can recruit multiple copies of antibodies. Through attaching transcriptional factors on the antibodies, the SunTag dCas9 activating complex amplifies its recruitment of transcriptional factors. In order to guide the dCas9 protein to its target gene, the dCas9 SunTag system uses sgRNA.
Tanenbaum et al.(2014) are credited for creating the dCas9 SunTag system. For the antibodies, they employed GCN4 antibodies which was bound to transcriptional factor VP64. In order to transport the antibodies to the nuclei of the cells, they attached NLS tag. To confirm the nuclear localization of the antibodies, sfGFP was used for visualization purpose. Therefore, the GCN4-sfGFP-NLS-VP64 protein was developed to be interact with dCas SunTag system. The antibodies successfully bound to SunTag polypeptides and activated target CXCR4 gene in K562 cell lines.[9] Comparing with the dCas9-VP64 activation complex, they were able to increase the CXCR4 gene expression 5-25 times greater in K562 cell lines. Not only was there a greater CXCR4 protein overexpression but also CXCR4 proteins were active to further travel on the transwell migration assay. Thus, the dCas9-SunTag system can be used to activate genes that are present latently such as virus genes.
3. Applications
The dCas9 activation system allows a desired gene or multiple genes in the same cell to be expressed. It is possible to study genes involved in a certain process using a genome wide screen that involves activating expression of genes. Examining which sgRNAs yield a phenotype suggests which genes are involved in a specific pathway. The dCas9 activation system can be used to control exactly which cells are activated and at what time activation occurs. dCas9 constructs have been made that turn on a dCas9-activator fusion protein in the presence of light or chemicals. Cells can also be reprogrammed or differentiated from one cell type into another by increasing the expression of certain genes important for the formation or maintenance of a cell type.[10]
3.1. Greater Control over Gene Expression
One research group used a system in which dCas9 was fused to a particular domain, C1B1. When blue light is shined on the cell, the Cry2 domain binds to C1B1. The Cry2 domain is fused to a transcriptional activator, so blue light targets the activator to the spot where dCas9 is bound. The use of light allows a great deal of control over when the targeted gene is activated. Removing the light from the cell results in only dCas9 remaining at the target gene, so expression is not increased. In this way, the system is reversible.[11] A similar system was developed using chemical control. In this system, dCas9 recruits an MS2 fusion protein that contains the domain FKBP. In the presence of the chemical RAP, an FRB domain fused to a chromatin modifying complex binds to FKBP. Whenever RAP is added to the cells, a specific chromatin modifier complex can be targeted to the gene. That allows scientists to examine how specific chromatin modifications affect the expression of a gene.[12] The dCAs9-VPR system is used as an activator by targeting it to the promoter of a gene upstream of the coding region. A study used various sgRNAs to target different portions of the gene, finding that the dCas9-VPR activator can act as an activator or a repressor, depending on the location it binds. In a cell, sgRNAs targeting the promoter could allow dCas9-VPR to increase expression, while sgRNAs targeting the coding region of the gene result in dCas9-VPR decreasing expression.[13]
3.2. Genome Wide Activation
The versatility of sgRNAs allows dCas9 activators to increase the expression of any gene within an organism's genome. That could be used to increase expression of a protein coding gene or a transcribed RNA. A paper demonstrated that genome wide activation could be used to determine which proteins are involved in mediated resistance to a specific drug.[5] Another paper used genome wide activation of long, noncoding RNAs and observed that increasing the expression of certain long noncoding RNAs conferred resistance to the drug vemurafenib.[14] In both cases, the cells that survive the drug could be studied to determine which sgRNAs they contain. That allows researchers to determine which gene was activated in each surviving cell, which suggests which genes are important for resistance to that drug.
3.3. Use in Organisms
A dCas9 fusion with VP64, p65, and HSF1 (heat shock factor 1) allowed researchers to target genes in Arabidopsis thaliana and increase transcription to a similar level as when the gene itself is inserted into the plant's genome. For one of the two genes tested, the dCas9 activator changes the number and size of leaves and made the plants better able to handle drought. The authors conclude that the dCas9 activator can create phenotypes in plants that are similar to those observed when a transgene is inserted for overexpression.[15] Researchers have used multiple guide RNAs to target dCas9 activation system to multiple genes in a specific mouse strain in which dCas9 can be turned on in specific cell lines using the Cre recombinase system. Scientists used the targeting and increased expression of several genes to examine the processes involved in regeneration and carcinomas of the liver.[16]
The content is sourced from: https://handwiki.org/wiki/Biology:DCas9_activation_system
References
- "CRISPR/Cas9 Guide". Addgene. https://www.addgene.org/crispr/guide/.
- Ma, J. (August 2011). Transcriptional activators and activation mechanisms. Protein and Cell, 2(11), 879-888.
- "RNA-guided gene activation by CRISPR-Cas9-based transcription factors". Nature Methods 10 (10): 973–6. October 2013. doi:10.1038/nmeth.2600. PMID 23892895. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=3911785
- "CRISPR RNA-guided activation of endogenous human genes". Nature Methods 10 (10): 977–9. October 2013. doi:10.1038/nmeth.2598. PMID 23892898. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=3794058
- "Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex". Nature 517 (7536): 583–8. January 2015. doi:10.1038/nature14136. PMID 25494202. Bibcode: 2015Natur.517..583K. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=4420636
- "Highly efficient Cas9-mediated transcriptional programming". Nature Methods 12 (4): 326–8. April 2015. doi:10.1038/nmeth.3312. PMID 25730490. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=4393883
- "Comparison of Cas9 activators in multiple species". Nature Methods 13 (7): 563–567. July 2016. doi:10.1038/nmeth.3871. PMID 27214048. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=4927356
- "CRISPR/gRNA-directed synergistic activation mediator (SAM) induces specific, persistent and robust reactivation of the HIV-1 latent reservoirs". Scientific Reports 5 (1): 16277. November 2015. doi:10.1038/srep16277. PMID 26538064. Bibcode: 2015NatSR...516277Z. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=4633726
- "A protein-tagging system for signal amplification in gene expression and fluorescence imaging". Cell 159 (3): 635–46. October 2014. doi:10.1016/j.cell.2014.09.039. PMID 25307933. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=4252608
- "Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation". Nature Reviews. Molecular Cell Biology 17 (1): 5–15. January 2016. doi:10.1038/nrm.2015.2. PMID 26670017. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=4922510
- "CRISPR-Cas9-based photoactivatable transcription system". Chemistry & Biology 22 (2): 169–74. February 2015. doi:10.1016/j.chembiol.2014.12.011. PMID 25619936. https://dx.doi.org/10.1016%2Fj.chembiol.2014.12.011
- "Rapid and reversible epigenome editing by endogenous chromatin regulators". Nature Communications 8 (1): 560. September 2017. doi:10.1038/s41467-017-00644-y. PMID 28916764. Bibcode: 2017NatCo...8..560B. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=5601922
- "Enabling Graded and Large-Scale Multiplex of Desired Genes Using a Dual-Mode dCas9 Activator in Saccharomyces cerevisiae". ACS Synthetic Biology 6 (10): 1931–1943. October 2017. doi:10.1021/acssynbio.7b00163. PMID 28700213. https://dx.doi.org/10.1021%2Facssynbio.7b00163
- "Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood". Nature 548 (7667): 343–346. August 2017. doi:10.1038/nature23451. PMID 28792927. Bibcode: 2017Natur.548..343J. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=5706657
- "RNA-guided transcriptional activation via CRISPR/dCas9 mimics overexpression phenotypes in Arabidopsis". PLOS One 12 (6): e0179410. 2017. doi:10.1371/journal.pone.0179410. PMID 28622347. Bibcode: 2017PLoSO..1279410P. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=5473554
- "Combinatorial genetics in liver repopulation and carcinogenesis with a in vivo CRISPR activation platform". Hepatology 68 (2): 663–676. August 2018. doi:10.1002/hep.29626. PMID 29091290. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=5930141
