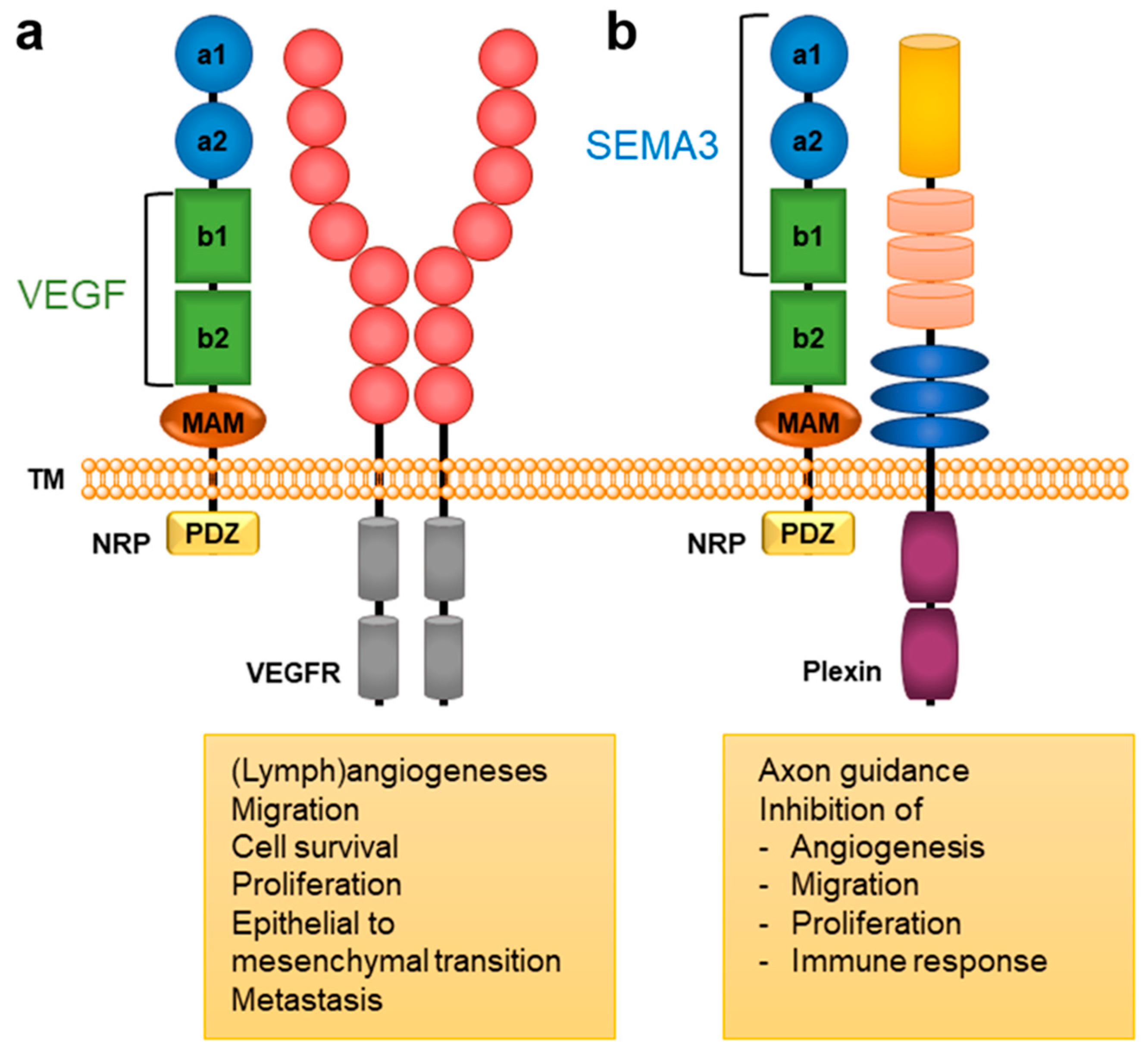
Neuropilin 1 (NRP1) represents one of the two homologous neuropilins (NRP, splice variants of neuropilin 2 are the other) found in all vertebrates. It forms a transmembrane glycoprotein distributed in many human body tissues as a (co)receptor for a variety of different ligands.
- neuropilins
- computer-aided drug design
- in silico drug design
1. Introduction
| Endogenous Ligand | Preferences for NRP Binding | Reference | |
|---|---|---|---|
| NRP1 | NRP2 | ||
| SEMA | SEMA3A SEMA3B SEMA3C SEMA3D SEMA3F |
SEMA3B SEMA3C SEMA3D SEMA3F SEMA3G |
[2][9] |
| VEGF | VEGF-A VEGF-A165 VEGF-A189 VEGF-B VEGF-C VEGF-D VEGF-E PIGF |
VEGF-A VEGF-A145 VEGF-A165 VEGF-C VEGF-D PIGF |
[3][9] |
| FGF | FGF-1 FGF-2 FGF-4 FGF-7 |
FGF-2 | [18] |
| HGF | x | x | [19] |
| PDGF | PDGF-BB PDGF-C PDGF-D |
PDGF-BB | [3][20] |
| TGF-β | TGF-β1 | TGF-β1 | [21][22] |
| miRNAs | x | x | [2][23] |
2. NRP Binding of Endogenous Ligands
3. Neuropilin-Related Pathology
3.1. Pain
3.2. Viral Entry
3.3. Cardiovascular Diseases
3.4. Diabetes
3.5. Cancer
|
Cancer type |
NRP1 |
NRP2 |
Reference |
|
Leukaemia |
x |
|
[86-91] |
|
Breast cancer |
x |
x |
[92-102] |
|
Carcinoma |
x |
x |
[103-116] |
|
Colon & Colorectal cancer |
x |
x |
[117-126] |
|
Gastric cancer |
x |
|
[127-138] |
|
Lung cancer |
x |
x |
[139-149] |
|
Pancreatic cancer |
x |
|
[150-154] |
|
Prostate cancer |
x |
x |
[155-157] |
|
Melanoma |
x |
x |
[158-1161] |
|
Glioma |
x |
|
[162-1168] |
|
Liver cancer |
x |
|
[169] |
|
Mammary stem cells cancer |
x |
|
[170,171] |
|
Esophageal cancer |
x |
|
[172,173] |
|
Stem cell cancer |
x |
|
[174,175] |
|
Thyroid cancer |
|
x |
[176,177] |
|
Multiple myeloma |
x |
|
[178] |
|
Lymphoma |
x |
|
[179] |
|
Bladder cancer |
|
x |
[180,181] |
|
Tongue cancer |
|
x |
[182] |
|
Cervical cancer |
x |
|
[183,184] |
|
Gallbladder cancer |
x |
|
[185] |
|
Endometrium cancer |
x |
x |
[186,187] |
This entry is adapted from the peer-reviewed paper 10.3390/ijms23158402
References
- Pellet-Many, C.; Frankel, P.; Jia, H.; Zachary, I. Neuropilins: Structure, function and role in disease. Biochem. J. 2008, 411, 211–226.
- Roy, S.; Bag, A.K.; Singh, R.K.; Talmadge, J.E.; Batra, S.K.; Datta, K. Multifaceted role of neuropilins in the immune system: Potential targets for immunotherapy. Front. Immunol. 2017, 8, 1228.
- Niland, S.; Eble, J.A. Neuropilin: Handyman and power broker in the tumor microenvironment. Adv. Exp. Med. Biol. 2020, 1223, 31–67.
- Carmeliet, P.; Tessier-Lavigne, M. Common mechanisms of nerve and blood vessel wiring. Nature 2005, 436, 193–200.
- Mayi, B.S.; Leibowitz, J.A.; Woods, A.T.; Ammon, K.A.; Liu, A.E.; Raja, A. The role of Neuropilin-1 in COVID-19. PLoS Pathog. 2021, 17, e1009153.
- Yelland, T.; Djordjevic, S. Crystal structure of the neuropilin-1 MAM domain: Completing the neuropilin-1 ectodomain picture. Structure 2016, 24, 2008–2015.
- Nakamura, F.; Goshima, Y. Structural and functional relation of neuropilins. Neuropilin: From Nervous System to Vascular and Tumor Biol. Adv. Exp. Med. Bio 2002, 515, 55–69.
- Nasarre, P.; Gemmill, R.M.; Drabkin, H.A. The emerging role of class-3 semaphorins and their neuropilin receptors in oncology. Onco Targets Ther. 2014, 7, 1663.
- Prud’homme, G.J.; Glinka, Y. Neuropilins are multifunctional coreceptors involved in tumor initiation, growth, metastasis and immunity. Oncotarget 2002, 3, 921.
- He, Z.; Tessier-Lavigne, M. Neuropilin is a receptor for the axonal chemorepellent Semaphorin III. Cell 1997, 90, 739–751.
- Kolodkin, A.L.; Levengood, D.V.; Rowe, E.G.; Tai, Y.T.; Giger, R.J.; Ginty, D.D. Neuropilin is a semaphorin III receptor. Cell 1997, 90, 753–762.
- Soker, S.; Takashima, S.; Miao, H.Q.; Neufeld, G.; Klagsbrun, M. Neuropilin-1 is expressed by endothelial and tumor cells as an isoform specific receptor for vascular endothelial growth factor. Cell 1998, 92, 735–745.
- Zhao, L.; Chen, H.; Lu, L.; Wang, L.; Zhang, X.; Guo, X. New insights into the role of coreceptor neuropilins in tumour angiogenesis and lymphangiogenesis and targeted therapy strategies. J. Drug Target. 2021, 29, 155–167.
- Takahashi, T.; Fournier, A.; Nakamura, F.; Wang, L.H.; Murakami, Y.; Kalb, R.G.; Fujisawa, H.; Strittmatter, S.M. Plexin-neuropilin-1 complexes form functional semaphorin-3A receptors. Cell 1999, 99, 59–69.
- Tamagnone, L.; Artigiani, S.; Chen, H.; He, Z.; Ming, G.L.; Song, H.J.; Chedotal, A.; Winberg, M.L.; Goodman, C.S.; Poo, M.; et al. Plexins are a large family of receptors for transmembrane, secreted, and GPI-anchored semaphorins in vertebrates. Cell 1999, 99, 71–80.
- Meyer, L.A.; Fritz, J.; Pierdant-Mancera, M.; Bagnard, D. Current drug design to target the Semaphorin/Neuropilin/Plexin complexes. Cell Adhes. Migr. 2016, 10, 700–708.
- Toledano, S.; Nir-Zvi, I.; Engelman, R.; Kessler, O.; Neufeld, G. Class-3 semaphorins and their receptors: Potent multifunctional modulators of tumor progression. Int. J. Mol. Sci. 2019, 20, 556.
- West, D.C.; Rees, C.G.; Duchesne, L.; Patey, S.J.; Terry, C.J.; Turnbull, J.E.; Delehedde, M.; Heegaard, C.W.; Allain, F.; Vanpouille, C.; et al. Interactions of multiple heparin binding growth factors with neuropilin-1 and potentiation of the activity of fibroblast growth factor-2. J. Biol. Chem. 2005, 280, 13457–13464.
- Sulpice, E.; Plouet, J.; Bergé, M.; Allanic, D.; Tobelem, G.; Merkulova-Rainon, T. Neuropilin-1 and neuropilin-2 act as coreceptors, potentiating proangiogenic activity. Blood 2008, 111, 2036–2045.
- Pellet-Many, C.; Mehta, V.; Fields, L.; Mahmoud, M.; Lowe, V.; Evans, I.; Ruivo, J.; Zachary, I. Neuropilins 1 and 2 mediate neointimal hyperplasia and re-endothelialisation following arterial injury. Cardiovasc. Res. 2015, 108, 288–298.
- Harman, J.L.; Sayers, J.; Chapman, C.; Pellet-Many, C. Emerging Roles for Neuropilin-2 in Cardiovascular Disease. Int. J. Mol. Sci. 2020, 21, 5154.
- Wittmann, P.; Grubinger, M.; Gröger, C.; Huber, H.; Sieghart, W.; Peck-Radosavljevic, M.; Mikulits, W. Neuropilin-2 induced by transforming growth factor-β augments migration of hepatocellular carcinoma cells. BMC Cancer 2015, 15, 909.
- Do, Y.; Cho, J.G.; Park, J.Y.; Oh, S.; Park, D.; Yoo, K.H.; Lee, M.S.; Kwon, B.S.; Kim, J.; Yang, Y. MiR-146a Regulates Migration and Invasion by Targeting NRP2 in Circulating-Tumor Cell Mimicking Suspension Cells. Genes 2020, 12, 45.
- Peng, K.; Bai, Y.; Zhu, Q.; Hu, B.; Xu, Y. Targeting VEGF–neuropilin interactions: A promising antitumor strategy. Drug Discov. 2019, 24, 656–664.
- Simons, M.; Gordon, E.; Claesson-Welsh, L. Mechanisms and regulation of endothelial VEGF receptor signalling. Nat. Rev. Mol. Cell Biol. 2016, 17, 611–625.
- Teesalu, T.; Sugahara, K.N.; Kotamraju, V.R.; Ruoslahti, E. C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc. Natl. Acad. Sci. USA 2019, 106, 16157–16162.
- Miao, H.Q.; Klagsbrun, M. Neuropilin is a mediator of angiogenesis. Cancer Metastasis Rev. 2000, 19, 29–37.
- Lampropoulou, A.; Ruhrberg, C. Neuropilin regulation of angiogenesis. Biochem. Soc. Trans. 2014, 42, 1623–1628.
- Staton, C.A.; Kumar, I.; Reed, M.W.R.; Brown, N.J. Neuropilins in physiological and pathological angiogenesis. J. Pathol. 2007, 212, 237–248.
- Mei, B.; Chen, J.; Yang, N.; Peng, Y. The regulatory mechanism and biological significance of the Snail-miR590-VEGFR-NRP1 axis in the angiogenesis, growth and metastasis of gastric cancer. Cell Death Dis. 2020, 11, 241.
- Hu, C.; Jiang, X. Role of NRP-1 in VEGF-VEGFR2-independent tumorigenesis. Target. Oncol. 2016, 11, 501–505.
- Alghamdi, A.A.; Benwell, C.J.; Atkinson, S.J.; Lambert, J.; Johnson, R.T.; Robinson, S.D. NRP2 as an emerging angiogenic player; promoting endothelial cell adhesion and migration by regulating recycling of α5 integrin. Front. Cell Dev. Biol. 2020, 8, 395.
- Iragavarapu-Charyulu, V.; Wojcikiewicz, E.; Urdaneta, A. Semaphorins in angiogenesis and autoimmune diseases: Therapeutic targets? Front. Immunol. 2020, 11, 346.
- Caunt, M.; Mak, J.; Liang, W.C.; Stawicki, S.; Pan, Q.; Tong, R.K.; Plowman, G.; Kowalski, J.; Ho, C.; Reslan, H.B.; et al. Supplemental Data Blocking Neuropilin-2 Function Inhibits Tumor Cell Metastasis. Cancer Cell 2008, 13, 331–342.
- Jarvis, A.; Allerston, C.K.; Jia, H.; Herzog, B.; Garza-Garcia, A.; Winfield, N.; Ellard, K.; Aqil, R.; Lynch, R.; Chapman, C.; et al. Small molecule inhibitors of the neuropilin-1 vascular endothelial growth factor A (VEGF-A) interaction. J. Med. Chem. 2010, 53, 2215–2226.
- Mota, F.; Fotinou, C.; Rana, R.R.; Chan, A.; Yelland, T.; Arooz, M.T.; O’Leary, A.P.; Hutton, J.; Frankel, P.; Zachary, I.; et al. Architecture and hydration of the arginine-binding site of neuropilin-1. FEBS J. 2018, 285, 1290–1304.
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.E.; Williamson, M.K.; Antón-Plágaro, C.; Shoemark, D.K.; Simón-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865.
- Parker, M.W.; Xu, P.; Li, X.; Vander Kooi, C.W. Structural basis for selective vascular endothelial growth factor-A (VEGF-A) binding to neuropilin-1. J. Biol. Chem. 2012, 287, 11082–11089.
- Llorián-Salvador, M.; González-Rodríguez, S. Painful understanding of VEGF. Front. Pharmacol. 2018, 9, 1267.
- Perez-Miller, S.; Patek, M.; Moutal, A.; Duran, P.; Cabel, C.R.; Thorne, C.A.; Khanna, R. Novel compounds targeting neuropilin receptor 1 with potential to interfere with SARS-CoV-2 virus entry. ACS Chem. Neurosci. 2021, 12, 1299–1312.
- Hulse, R.P. Role of VEGF-A in chronic pain. Oncotarget 2017, 8, 10775.
- Wang, H.B.; Zhang, H.; Zhang, J.P.; Li, Y.; Zhao, B.; Feng, G.K.; Zeng, M.S. Neuropilin 1 is an entry factor that promotes EBV infection of nasopharyngeal epithelial cells. Nat. Commun. 2015, 6, 6240.
- Ghez, D.; Lepelletier, Y.; Lambert, S.; Fourneau, J.M.; Blot, V.; Janvier, S.; Hermine, O. Neuropilin-1 is involved in human T-cell lymphotropic virus type 1 entry. J. Virol. 2006, 80, 6844.
- Lambert, S.; Bouttier, M.; Vassy, R.; Seigneuret, M.; Petrow-Sadowski, C.; Janvier, S.; Pique, C. HTLV-1 uses HSPG and neuropilin-1 for entry by molecular mimicry of VEGF165. Blood 2009, 113, 5176–5185.
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860.
- Lane, R.K.; Guo, H.; Fisher, A.D.; Diep, J.; Lai, Z.; Chen, Y.; Kaiser, W.J. Necroptosis-based CRISPR knockout screen reveals Neuropilin-1 as a critical host factor for early stages of murine cytomegalovirus infection. Proc. Natl. Acad. Sci. USA 2020, 117, 20109–20116.
- Wang, H.C.; Huang, P.N.; Hung, H.C.; Tseng, S.N.; Chang, C.C.; Tsai, Y.R.; Hsu, J.T. Effect of a Neuropilin-1-Derived Virus Receptor Trap on Enterovirus A71 infection in vitro. Antimicrob. Agents Chemother. 2020, 65, e00695-20.
- Raaben, M.; Jae, L.T.; Herbert, A.S.; Kuehne, A.I.; Stubbs, S.H.; Chou, Y.Y.; Whelan, S.P. NRP2 and CD63 are host factors for Lujo virus cell entry. Cell Host Microbe 2017, 22, 688–696.
- Martinez-Martin, N.; Marcandalli, J.; Huang, C.S.; Arthur, C.P.; Perotti, M.; Foglierini, M.; Ciferri, C. An unbiased screen for human cytomegalovirus identifies neuropilin-2 as a central viral receptor. Cell 2018, 174, 1158–1171.
- Kusunoki, H.; Tanaka, T.; Kohno, T.; Matsuhashi, K.; Hosoda, K.; Wakamatsu, K.; Hamaguchi, I. A novel neuropilin-1–binding sequence in the human T-cell lymphotropic virus type 1 envelope glycoprotein. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 541–548.
- Gu, Y.Y.; Luo, B.; Li, C.Y.; Huang, L.S.; Chen, G.; Feng, Z.B.; Peng, Z.G. Expression and clinical significance of neuropilin-1 in Epstein-Barr virus-associated lymphomas. Cancer Biomark. 2019, 25, 259–273.
- Hwang, J.Y.; Sun, Y.; Carroll, C.R.; Usherwood, E.J. Neuropilin-1 regulates the secondary CD8 T cell response to virus infection. mSphere 2019, 4, e00221-19.
- Wang, Y.; Cao, Y.; Yamada, S.; Thirunavukkarasu, M.; Nin, V.; Joshi, M.; Rishi, M.T.; Bhattacharya, S.; Camacho-Pereira, J.; Sharma, A.K.; et al. Cardiomyopathy and Worsened Ischemic Heart Failure in SM22-α Cre-Mediated Neuropilin-1 Null Mice: Dysregulation of PGC1α and Mitochondrial Homeostasis. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 1401–1412.
- Kilari, S.; Wang, Y.; Singh, A.; Graham, R.P.; Iyer, V.; Thompson, S.M.; Torbenson, M.S.; Mukhopadhyay, D.; Misra, S. Neuropilin-1 deficiency in vascular smooth muscle cells is associated with hereditary hemorrhagic telangiectasia arteriovenous malformations. JCI Insight 2022, 7, e155565.
- Matilla, L.; Arrieta, V.; Jover, E.; Garcia-Peña, A.; Martinez-Martinez, E.; Sadaba, R.; Alvarez, V.; Navarro, A.; Fernandez-Celis, A.; Gainza, A.; et al. Soluble St2 Induces Cardiac Fibroblast Activation and Collagen Synthesis via Neuropilin-1. Cells 2020, 9, 1667.
- Li, B.; Yang, J.; Zhao, F.; Zhi, L.; Wang, X.; Liu, L.; Bi, Z.; Zhao, Y. Prevalence and impact of cardiovascular metabolic diseases on COVID-19 in China. Clin. Res. Cardiol. 2020, 109, 531–538.
- Sahni, S.; Gupta, G.; Sarda, R.; Pandey, S.; Pandey, R.M.; Sinha, S. Impact of metabolic and cardiovascular disease on COVID-19 mortality: A systematic review and meta-analysis. Diabetes Metab. Syndr. 2021, 15, 102308.
- Chen, L.; Li, X.; Chen, M.; Feng, Y.; Xiong, C. The ACE2 expression in human heart indicates new potential mechanism of heart injury among patients infected with SARS-CoV-2. Cardiovasc. Res. 2020, 116, 1097–1100.
- Nicin, L.; Abplanalp, W.T.; Mellentin, H.; Kattih, B.; Tombor, L.; John, D.; Schmitto, J.D.; Heineke, J.; Emrich, F.; Arsalan, M.; et al. Cell type-specific expression of the putative SARS-CoV-2 receptor ACE2 in human hearts. Eur. Heart J. 2020, 41, 1804–1806.
- Hikmet, F.; Méar, L.; Edvinsson, Å.; Micke, P.; Uhlén, M.; Lindskog, C. The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol. 2020, 16, e9610.
- Klagsbrun, M.; Takashima, S.; Mamluk, R. The role of neuropilin in vascular and tumor biology. In Neuropilin: From Nervous System to Vascular and Tumor Biology; Springer: Berlin/Heidelberg, Germany, 2002; pp. 33–48.
- Mourad, D.; Azar, N.S.; Azar, S.T. Diabetic nephropathy and COVID-19: The potential role of immune actors. Int. J. Mol. Sci. 2021, 22, 7762.
- Wang, J.; Wang, S.; Li, M.; Wu, D.; Liu, F.; Yang, R.; Ji, S.; Ji, A.; Li, Y. The neuropilin-1 inhibitor, ATWLPPR peptide, prevents experimental diabetes-induced retinal injury by preserving vascular integrity and decreasing oxidative stress. PLoS ONE 2015, 10, e0142571.
- Loeffler, I.; Rüster, C.; Franke, S.; Liebisch, M.; Wolf, G. Erythropoietin ameliorates podocyte injury in advanced diabetic nephropathy in the db/db mouse. Am. J. Physiol. Ren. Physiol. 2013, 305, F911–F918.
- Bondeva, T.; Wolf, G. Role of Neuropilin-1 in Diabetic Nephropathy. J. Clin. Med. 2015, 4, 1293–1311.
- International Agency for Research on Cancer. Available online: https://gco.iarc.fr/today/ (accessed on 24 June 2022).
- Roy, S.; Bag, A.K.; Dutta, S.; Polavaram, N.S.; Islam, R.; Schellenburg, S.; Banwait, J.; Guda, C.; Ran, S.; Hollingsworth, M.A.; et al. Macrophage-Derived Neuropilin-2 Exhibits Novel Tumor-Promoting Functions. Cancer Res. 2018, 78, 5600–5617.
- Diaz-Vera, J.; Palmer, S.; Hernandez-Fernaud, J.R.; Dornier, E.; Mitchell, L.E.; Macpherson, I.; Edwards, J.; Zanivan, S.; Norman, J.C. A Proteomic Approach to Identify Endosomal Cargoes Controlling Cancer Invasiveness. J. Cell Sci. 2017, 78, 5600–5617.
- Chen, D.S.; Mellman, I. Oncology Meets Immunology: The Cancer-Immunity Cycle. Immunity 2013, 39, 1–10.
