1. Introduction
In nature, plants are continuously challenged by a mix of environmental conditions, including temperature fluctuation, elevated atmospheric CO
2, drought, flooding, saline soil, a variety of pathogens and insect herbivores. To cope with the simultaneous exposure to combined external variations, plants have evolved inducible mechanisms to balance assimilates and signaling molecules between growth and defense. Such a trade-off is believed to be primarily due to resource restrictions [
1]. The regulation network relies on several common signaling pathways, including phytohormone balance, the production of reactive oxygen species (ROS), calcium signatures, the activation of kinase cascades, and sugar signals [
1,
2] to ensure robust defense response with minimum fitness cost. How plants integrate these inducible mechanisms to cope with combined biological and physical stimuli and to balance growth and defense remains largely unexplored.
Recent reports suggest that plant responses to combined stresses might not simply be additive of individual stresses [
3,
4]. Meta-analysis indicates that gene expression under multifactorial stress is not predictable from single stress exposures [
5,
6], arguing for investigating plant responses to different combinations of biotic and abiotic stresses. However, different laboratory conditions and variation in plant species, pathogens, treatment time, and strength lead to discrepancies between published works. Nevertheless, some independent reports do converge on common phytohormone signaling or central metabolic pathways, highlighting the significance of these important hubs, which form the foundation for further exploration. In general, phytohormones such as salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) are considered major regulators of plant defense responses. Conventionally, SA signaling is induced by and involved in defense against biotrophic and hemi-biotrophic pathogens and viral infection, as well as for establishing systemic acquired resistance [
7,
8]. On the other hand, JA/ET signaling is activated by necrotrophic pathogens and insect herbivores [
9]. For combined stresses, the signature and corresponding transcriptional changes might be difficult to evaluate and will be a future challenge. In this review, we summarize current updates on the impact of several environmental factors on plant immunity, focusing on their roles in modulating the SA-dependent signaling pathway. This information might provide a framework to further explore the signaling network underlying the growth–defense trade-off (
Figure 1) and in the long run to translate laboratory findings to field application.
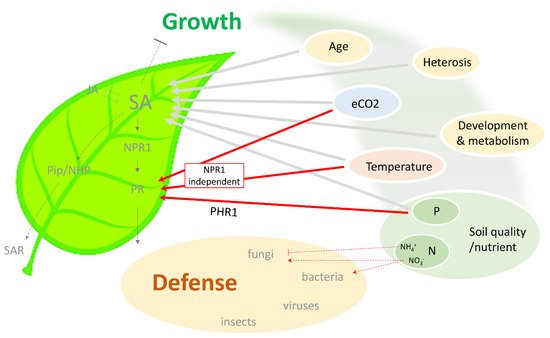
Figure 1. Schematic summary of the connection between different environmental conditions and SA-dependent immunity. Plant growth depends on multiple environmental factors, including temperature, atmospheric CO2, and soil quality (nutrient status). These factors have also been found to modulate SA level and SA-dependent defense response against various pathogens. Notably, an NPR1-independent pathway exists in external factor-mediated protection, which potentially bypasses SA and the associated negative effect on plant growth. SA, salicylic acid; JA, jasmonic acid; NPR1, nonexpressor of pathoegenesis-related genes 1; PR, pathogenesis-related; Pip, pipecolic acid; NHP, N-hydroxy-pipecolic acid; SAR, systemic acquired resistance; eCO2, elevated CO2; P, phosphorus; N, nitrogen; PHR1, phosphate starvation response 1.
2. SA Biosynthesis, Perception, and Signaling in Defense and Growth
SA is an essential phytohormone for basal immunity and systemic acquired resistance (SAR) against biotrophic and hemi-biotrophic pathogens. The biosynthesis of SA has been thoroughly described in excellent reviews [
7,
8]. Briefly, SA is synthesized by two independent pathways using chorismate or phenylpropanoid as the precursors. The isochorismate synthase (ICS) pathway is the major process that mediates SA induction upon pathogen infection. In the Arabidopsis (
Arabidopsis thaliana) genome, two isochrismate synthase genes (
ICS1 and
ICS2) encode the enzymes that convert chorismate to isochrismate (IC) for initiating SA production in the chloroplast. Mutation of
ICS1 (
ics1, or alternatively
SA-deficient 2,
sid2 mutant) hampers up to 90% SA induction upon pathogen infection, indicating that
ICS1 plays a major role over
ICS2 in defense. IC in the chloroplast is transported to the cytosol by the MATE transporter enhanced disease susceptibility 5 (EDS5) [
10,
11,
12]. Finally, avrPphB susceptible 3 (PBS3) catalyzes the conjugation of IC to glutamate (Glu), producing IC9-Glu, which spontaneously breaks down in the cytosol to produce SA [
10,
13].
ICS1,
EDS5, and
PBS3 are strongly induced upon pathogen infection. Glucose conjugation of SA produces SA glucoside (SAG), which is actively transported from the cytosol into the vacuole as an inactive storage form. Although the ICS pathway is seemingly important for SA production, the alternative pathway—the phenylalanine ammonia lyase (PAL) pathway—is also indispensable. Quadruple mutation of Arabidopsis
PAL genes showed 75% SA reduction at rest and 50% SA reduction upon pathogen infection [
14]. The large reduction of pathogen-induced SA level when either the ICS or PAL pathway is disrupted is puzzling. How ICS and PAL pathways interplay to maintain the SA pool remains to be established.
Identification of cellular components for SA perception has mainly been carried out by forward genetic screening. To date, the most well-documented SA binding proteins are the nonexpressor of PR genes (NPRs), including NPR1, NPR3 and NPR4, which serve as bona fide SA receptors [
15]. Recombinant NPR1 binds to SA with a dissociation constant (Kd) range from 140 to 220 nM in vitro [
15,
16]. NPR1 is the master regulator for SA-induced PR gene expression and, hence, resistance against pathogens. Mutation of
NPR1 disables
PR gene expression and response to various SAR-inducing treatments and promotes susceptibility to infections [
17]. NPR1 protein consists of an N-terminal BTB/POZ domain, a central Ankyrin repeat (ANK) region, and a C-terminal transactivation domain [
17,
18]. Interestingly, NPR1 does not bear a DNA-binding domain. Therefore, NPR1 is considered a transcriptional co-activator that depends on physical interaction with a partner transcription factor, such as WRKYs and TGAs, for activating downstream SA-responsive genes.
SA also plays an important role in plant growth and development [
19]. In general, SA over-accumulation negatively affects plant growth. Exogenous SA application inhibits vegetative growth in soybean (
Glycine max) [
20], wheat (
Triticum aestivum) [
21], maize (
Zea mays) [
22], and chamomile (
Matricaria chamomilla) [
23]. Many autoimmune mutants have been reported, and many of them exhibit SA over-accumulation and dwarfism [
24]. Notably, the growth inhibition in these autoimmune mutants can be rescued by blocking SA biosynthesis or SA signaling, reiterating the negative effect of SA on plant growth. Thus, mutation of SA biosynthetic enzyme (
sid2 mutant) or disrupting SA accumulation (by introducing
nahG transgene) leads to increased biomass and seed yield [
25].
3. Environmental Conditions Modulate SA Biosynthesis and SA Signaling
3.1. Temperature
Temperature sensitivity in plant disease resistance was first recognized in the 1900s [
26,
27]. The impact of elevated (and low) temperature on plant disease resistance was then tested in many plant–pathogen systems. In general, there seems to be a negative correlation between elevated temperature and disease resistance in plants.
Early studies showed that low temperature promoted the accumulation of SA and SAG in Arabidopsis, accompanied by upregulation of
ICS1,
CBP60g, and
SARD1 transcripts [
28]. The cold-stress-induced bacterial resistance phenotype was eventually confirmed by challenging Arabidopsis with the bacterial pathogen
Pseudomonas syringae pv.
tomato strain DC3000 (
Pst DC3000) [
29,
30]. Bacteria titer was significantly reduced using 4 °C [
29] or 16 °C [
30] treatment conditions. Notably, an intact SA signaling pathway is required for cold-stress-induced resistance.
Sid2 mutation [
30] and
nahG transgene [
29] abolished cold-stress-induced protection. Interestingly, mutation of
NPR1 only partially abolished cold-stress-induced protection, indicating that an NPR1-independent pathway exists.
On the other hand, elevated temperature promotes virus susceptibility in Arabidopsis [
3], tomato (
Solanum lycopersicum) [
31,
32], and potato (
Solanum tuberosum) [
33]. SA subverts heat-stress-induced viral sensitivity in potato cultivars. In
S. tuberosum L. cv. Gala,
StPR1-b gene expression was rapidly induced upon potato virus Y (PVY) infection at both 22 °C and 28 °C. However, in
S. tuberosum L. cv. Chicago,
StPR1-b gene expression was only induced at 22 °C but not 28 °C. Consistently, PVY RNA accumulation, as determined by qRT-PCR, revealed that Gala was more resistant, while the viral load in Chicago was up to 20-fold higher [
33]. In addition,
N gene conferred gene-for-gene resistance to the viral pathogen tobacco mosaic virus (TMV). Two independent studies confirmed that the hypersensitive response triggered by TMV was heat-dependent in tomato (
S. lycopersicum) [
31] and tobacco (
Nicotiana benthamiana) [
34]. Under elevated temperature, N protein showed reduced nuclear accumulation, presumably due to conformational change [
35]. Similarly, heat-dependent suppression of disease resistance has also been reported in plant–bacteria interactions. At 28 °C, Arabidopsis plants showed more severe disease symptoms and increased bacterial growth when challenged with virulent
Pst DC3000 [
34]. However,
sid2 mutation and
nahG transgenic plants retained temperature sensitivity, indicating that the involvement of SA signaling was minimal in this case study [
34].
3.2. Atmospheric CO2
The other important variant as a result of global climate change is elevated atmospheric CO
2 (eCO
2), which can modulate the balance between hormone levels in many plant species [
36]. The impact of eCO
2 on biological process besides primary metabolism includes biotic stress responses, as has been revealed by transcriptomic studies in Arabidopsis [
37] and wheat (
T. aestivum) [
38]. Quantification of defense hormones further confirmed the observation. Elevated CO
2 induced SA accumulation in Arabidopsis [
39,
40], beans (
Phaseolus vulgaris) [
40], wheat (
T. aestivum) [
40], and tomato (
S. lycopersicum) [
41]. The associated induction of SA marker gene
PR1 was stronger under eCO
2 in different plant species [
39,
40,
41]. Consistently, the plants showed enhanced resistance against the biotrophic oomycete
Hyaloperonospora arabidopsidis [
39] and
Pst DC3000 [
40,
41]. For instance, an intact NPR1-dependent pathway was required for bacteria resistance in tomato [
41], highlighting the specificity of an SA-dependent pathway in immune signaling mediated by eCO
2. However, in Arabidopsis
sid2 and
npr1 mutants, eCO
2-induced resistance against oomycete was only partially affected [
39], underlying the involvement of an alternative pathway other than the priming of SA-dependent defense. On the other hand, the opposite effect of eCO
2 has also been reported in other studies using different plant species and pathogen. In maize (
Z. mays), eCO
2 did not significantly induce SA nor JA levels under unchallenged conditions [
42]. However, after infection with the fungal pathogen
Fusarium verticillioides, JA induction was abolished while SA level was repressed. As a result, eCO
2 negatively affected fungal resistance in maize [
42]. In wheat (cv.
Remus), eCO
2 was shown to enhance virulence of the fungal pathogen
Zymoseptoria tritici and hence promoted plant susceptibility [
43].
Intriguingly, the antagonism between SA and JA was not consistent between Arabidopsis and tomato. Under eCO
2, both SA and JA levels were significantly induced in Arabidopsis [
39]. The JA-inducible gene
VSP2 was upregulated by ~400-fold, and the plants were more resistant to the necrotrophic fungus
Plectosphaerella cucumerina [
39]. A subsequent report by Zhou et al. (2019) [
44] also detected upregulation of the JA marker gene
PDF1.2 under eCO
2, and enhanced resistance against the necrotrophic fungus
Botrytis cinerea. However, in the same study, eCO
2 suppressed SA-signaling and reduced plant resistance against
Pst DC3000, which was opposite to the observation by Williams et al. (2018) [
39]. On the other hand, JA level was not significantly affected by eCO2 in tomato, and the plants were more susceptible to
B. cinerea infection [
41]. The discrepancy is likely due to a different experimental setup, as eCO
2 significantly affects plant development and canopy density [
45,
46], along with differences in other variables such as plant growth conditions and the strength and duration of pathogen treatment.
3.3. Nutrient Status Modulates SA-Dependent Immunity
Nitrogen and phosphorus (P) are the two major mineral nutrients that determine plant growth and productivity. Both nutrients have been implicated in plant defense responses [
55,
56]. Although it is generally accepted that nutrient status influences disease in plants/crops, much of the findings are contradictory, limiting the application of nutrient/fertilization to facilitate disease control. Two hypotheses shape the studies in determining nutrient–immunity crosstalk. First, pathogen proliferation depends on nutrient availability in planta. Thus, nutrient-limited plants might be better defended. Second, changes in host secondary metabolites in response to external nutrient supply determine plant resistance.
Nitrogen is acquired by plants from the soil as nitrate or ammonium [
57], or via symbiotic association with nitrogen-fixing bacteria in legumes [
58]. Early study showed that disease susceptibility of tomato (
Solanum esculentum) depended on external nitrogen supply and was pathogen-specific. Increasing nitrogen (nitrate) supply to tomato promoted susceptibility against the bacterial pathogen
P. syringae and the powdery mildew
Oidium lycopersicum in tomato [
59]. This observation appears to fit the first hypothesis that pathogens rely on nutrient availability in the plant. Subsequent reports in rice (
Oryza sativa) [
60,
61] and wheat (cv. Arche and Récital) [
61] using high nitrogen (both nitrate and ammonia) also led to enhanced susceptibility against fungal blast. However, nitrogen supply did not affect plant resistance/susceptibility against the wilt agent
Fusarium oxysporum f.sp.
lycopersici [
59] nor against
B. cinerea [
62]. Intriguingly, it has been observed that nitrogen content in the apoplast of tomato leaf was further increased after infection [
63]. This observation was further supported by a subsequent study to investigate nitrogen management in tomato plants after infection by different pathogens and by chemical elicitation. Glutamine synthetase (GS1) and glutamate dehydrogenase (GDH) regulate nitrogen mobilization in tobacco. Both marker genes were induced by SA, viruses (Cucumber Mosaic Virus, Tobacco Etch Virus, and PVY), bacteria (
P. syringae pv.
syringae,
hrp mutant, and
P. syringae pv.
tabaci), and fungal elicitors (cryptogein and Onozuka R10) [
64]. Similarly, in common bean (
P. vulgaris), infection by fungus (
Colletotrichum lindemuthianum) also activated nitrogen mobilization (
GS1 mRNA) and SA signaling (
PAL3 mRNA) [
65]. Interestingly, fungus preferred nitrate as the nitrogen source over ammonium, as tomato plants supplied with ammonium rather than nitrate showed significant reduction in vascular wilt symptoms when infected with the fungal pathogen
Fusarium oxysporum [
66].
This entry is adapted from the peer-reviewed paper 10.3390/cells11192985