Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Poly ADP-ribosylation (PARylation) is a post-translational modification process. Following the discovery of PARP-1, numerous studies have demonstrated the role of PARylation in the DNA damage and repair responses for cellular stress and DNA damage.
- PARylation
- poly(ADP-ribose) polymerases (PARPs)
- ADP-ribose
1. Introduction
Since the discovery of protein post-translational modifications (PTMs), PTMs have been recognized as important cellular regulatory mechanisms, and uncontrolled PTMs can induce disease or cellular abnormalities [1][2]. A PTM is a covalent process that alters the function or characteristics of a protein via adding functional groups or through the proteolytic cleavage of regulatory subunits. There are several types of PTM, which are classified based on their modification process or unique characteristics such as methylation, phosphorylation, acetylation, ubiquitylation, SUMOylation, glycosylation, and PARylation. These PTMs regulate various essential biological processes, such as gene expression, enzyme activity, protein stability, aging, and metabolism [2][3][4][5]. Phosphorylation is one of the most common PTMs, and it regulates the activity of enzymes or the function of proteins [6].
Poly ADP-ribosylation (PARylation) is another common PTM in eukaryotes [7][8]. In the last four decades, people's understanding of the function of poly(ADP-ribose) polymerases (PARPs) and PARylation has greatly expanded since the discovery of PARP [7]. PARylation is classified by the number of units of ADP-ribose (PAR), as mono PARylation (MARylation), oligo PARylation, and poly PARylation [9]. PARylation is catalyzed by PARPs, comprising 17 protein families in humans and 16 members in mice [9]. PARPs have one or more additional domains required for their unique roles [10]. Among the PARPs, PARP-1, PARP-2, PARP-5a, and PARP-5b induce poly PARylation during the DNA damage response (DDR) and DNA repair [11]. Other PARPs mainly induce MARylation in the nucleus or cytoplasm, except for PARP-9 and PARP-13 [12].
PARP-1, belonging to the first PARPs family, is the most abundant. It is the founding member of PARPs for the synthesis of ADP-ribose using nicotinamide adenine dinucleotide (NAD+) as a substrate [13]. PARP-1 contains an N-terminal DNA-binding domain, nuclear localization signal, central automodification domain, and a C-terminal catalytic domain. PARP-1 is ubiquitously expressed in mammalian cells, and is localized in the nucleus. PARP-1 is activated by the DDR, and is responsible for the majority (~90%) of global PAR synthesis following DNA strand breakage. PARPs covalently combine with the poly(ADP-ribose) (PAR) unit on the carboxyl group of acidic residues, such as glutamate, aspartate, and/or lysine residues in the target protein. PAR polymers may alter chromatin structure or disrupt protein–protein or DNA interactions due to their negative charge [14]. A recent proteomics study indicated that many DDR and DNA repair proteins are PARylated during their respective processes [15]. Interestingly, PARP-1 and PARP-2 can auto-PARylate themselves.
Historically, studies on PARylation have mainly focused on the DDR and repair pathways. Under DNA damage conditions, PARPs move to the DNA damage site and auto-PARylate themselves. This phenomenon induces chromatin remodeling and recruits other DNA repair proteins. However, recent studies have suggested novel roles for PARylation in inflammation, metabolism, and cell death [16][17][18][19].
2. Role of PARylation in DNA Damage Response and Repair
PARP-1, a DNA-damage-sensing protein, can attach a negatively charged PAR to itself or target proteins. This process involves the consumption of large amounts of cellular NAD+ under DNA damage [20][21]. PARylation has multiple roles in the DDR and DNA repair pathways, including the repair of single-strand breaks (SSBs), double-strand breaks (DSBs), DNA replication forks, and chromatin structures (Figure 1) [20][22][23].
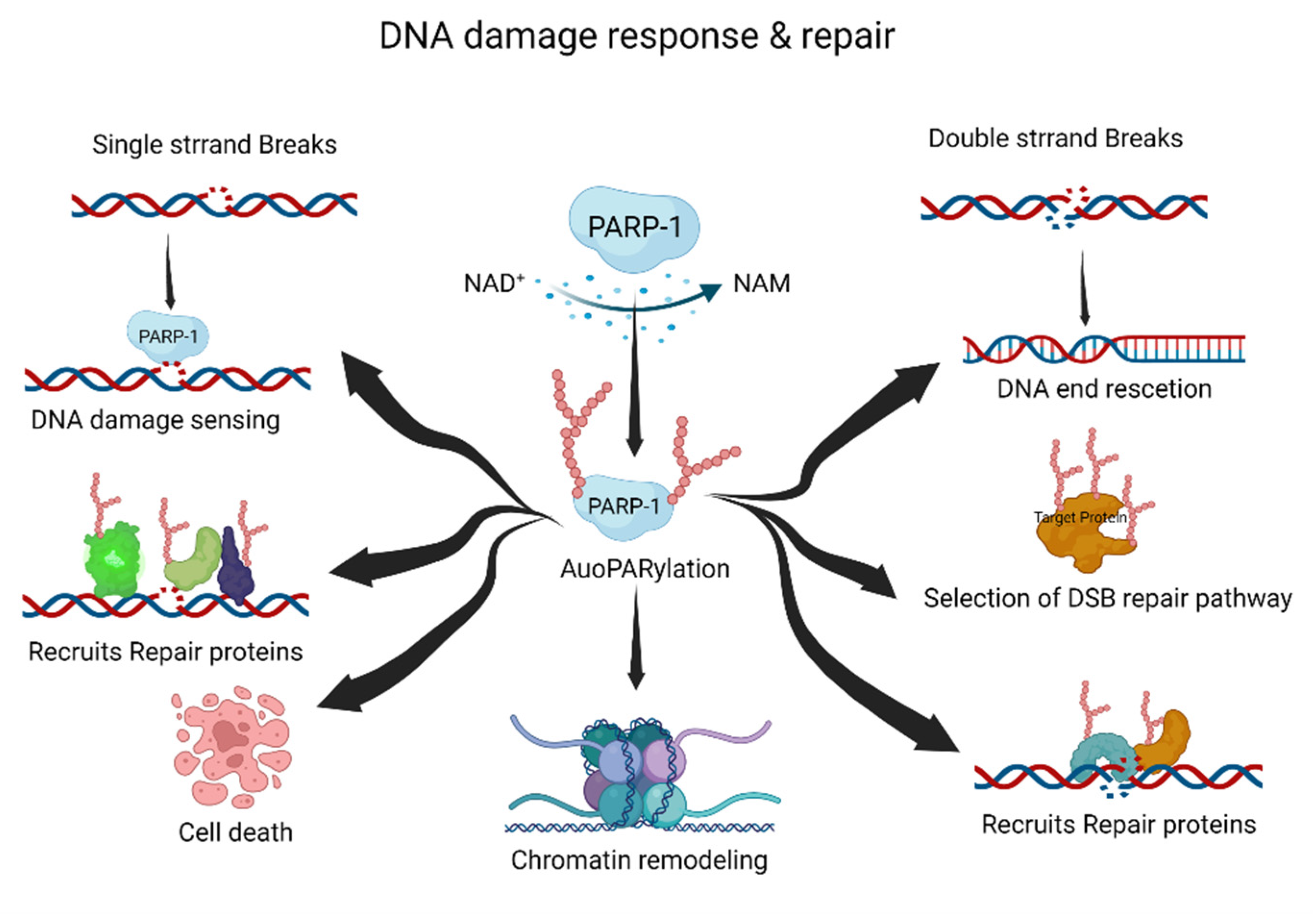
Figure 1. The roles of PARylation in DNA damage response and repair. PARylation acts as an initiator and/or selection effector of the DNA damage response and repair processes. NAD+, nicotinamide adenine dinucleotide; NAM, nicotinamide. This figure was created using BioRender.
Typically, PARylation is induced by cellular stress and DNA damage [24][25]. When DNA damage occurs, the DDR recruits PARP-1 to the DNA damage site. Recruited PARP-1 induces the auto-PARylation and PARylation of other proteins, including DNA repair and chromatin structure proteins. These processes are essential for the DDR and DNA damage repair pathways. The PARylated PARP-1 at the DNA damage site is composed of linear or branched repeats of ADP-ribose. Repeating ADP-ribose units are combined through ribose–ribose glycosidic bonds to generate linear repeats of ADP-ribose, and the linear chain is branched every 20–50 ADP-ribose repeat units [8]. DNA repair proteins and PARylated proteins bind to PAR on auto-PARylated PARP-1 through noncovalent interactions [26]. The linear or branched repeats of ADP-ribose recruit more DNA repair or PARylated proteins [11][24]. Interestingly, the branching or chain length of PARylation influences its functions in cellular physiology and stress responses [27].
SSBs are the most frequently generated form of DNA damage, and are caused by direct attacks from intracellular metabolites and/or spontaneous DNA decay. Therefore, the SSB repair pathway is one of the most important repair pathways [28]. PARP-1 rapidly detects SSBs and binds to the SSB region. Auto-PARylated PARP-1 then recruits X-ray repair cross-complementing protein 1 (XRCC1) for SSB repair. XRCC1 is the main effector of the SSB repair pathway, and acts as a scaffold for SSB repair proteins such as DNA ligase 3 and DNA polymerase β [29]. The mutation of XRCC1 in human and mouse models inhibits the SSB repair pathway, resulting in neuropathological defects with hyper-PARylation [30]. Hyper-PARylation in the unrepaired SSB repair pathway leads to the depletion of cellular NAD levels, which results in cell death. Nucleotide excision repair (NER) is also regulated by PARylation. The major NER repair pathway for bulky DNA lesions is induced by various mutagenic agents, such as ultraviolet (UV) irradiation [31][32]. The protein–protein complex of xeroderma pigmentosum C (XPC)–RAD23B recognizes UV damage and initiates the NER pathway [33]. XPC–RAD23B binds to PAR and PARylation of XPC–RAD23B modulates the recognition of UV damage sites [34]. DNA-damage-binding protein 2 (DDB2) stimulates histone PARylation, which induces nucleosome displacement and triggers the NER pathway [35]. Thus, PARylation plays an essential role in NER initiation and efficiency.
DSBs can be induced by various DNA-damaging agents, such as ionizing irradiation, chemicals, DNA replication, and DNA repair processes [36]. Homologous recombination (HR) and non-homologous end joining (NHEJ) are the major DSB repair pathways. Pathway selection is determined by the cell cycle stage and chromatin condition of DSBs. PARP-1 recognizes DSBs and induces PARylation, leading to the recruitment of DSB repair proteins [21][37]. Meiotic recombination 11 (MRE11) is recruited at the DSB site by PARylation, and is involved in DNA end resection and HR selection [38]. The function of breast cancer type 1 (BRCA1) is regulated by PARylation [39][40]. BRCA1 is recruited by PARylation at the DSB site, and PARylated BRCA1 regulates DNA recombination [41]. Furthermore, PARylation inhibits hyper-resected DNA DSB. The inhibition of PARylation induces hyper-resected DNA DSB due to the PARylation-mediated recruitment of Ku and p53-binding protein 1 (p53BP1) to DNA damage sites [37]. The role of PARylation in NHEJ is not as well explored as that in HR; however, some functions have been proposed. PARylation may enhance the recruitment of NHEJ repair proteins at DSB sites and promote NHEJ [42]. Therefore, PARylation is important for the initiation and accuracy of DSB repair.
Multiple cellular processes need to be coordinated during DNA damage repair. To regulate cellular processes such as transcription and replication, recruited chromatin remodelers control chromatin relaxation or condensation through PARylation [14][23][43][44][45]. PARP-1 and PARylation provide a scaffold for the recruitment of various DNA repair proteins and chromatin remodelers. PARylation of histone core proteins, including H2A, H2B, H3, and H4, may induce nucleosome disassembly, resulting in chromatin relaxation [45]. Proteins are then recruited by PARylation at the DNA damage site [23]. For example, PARylation is amplified in liver cancer 1 (ALC1/CHD1L) at the nucleosome disassembly site, and ALC1/CHD1L enhances the accessibility of DNA repair proteins to DNA damage sites [46]. Transcription repression factors are recruited via PARylation. The nucleosome remodeling and deacetylase complex, chromodomain helicase DNA-binding protein 4, and metastasis-associated 1, are recruited by PARylation [47]. PARylation removes nascent RNA and elongates RNA polymerase at the DNA damage site. Furthermore, PARylated PARP-1 binds to linker DNA, which induces chromatin condensation [23].
Following chromatin remodeling and the recruitment of DNA repair proteins to damaged sites, PAR is quickly released from the chromatin. Rapid PAR degradation is essential for cellular homeostasis and genomic stability. Hyper-PARylation or accumulation of PAR in chromatin induces genomic instability and/or cell death [48]. PARylated PARP-1 dissociates PARP-1 from the DNA damage site [49]. Catabolic enzymes such as poly(ADP-ribose) glycohydrolase (PARG), ADP-ribosyl hydrolase 3 (ADH3), and ADP-ribosyl protein lyase degrade PAR in the chromatin or nucleus [50][51][52]. These phenomena suggest that PARylation induces chromatin remodeling to increase the efficiency and accuracy of DNA repair in the DDR and DNA damage repair pathways.
This entry is adapted from the peer-reviewed paper 10.3390/ijms23179826
References
- Xie, H.; Vucetic, S.; Iakoucheva, L.M.; Oldfield, C.J.; Dunker, A.K.; Obradovic, Z.; Uversky, V.N. Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins. J. Proteome Res. 2007, 6, 1917–1932.
- Duan, G.; Walther, D. The roles of post-translational modifications in the context of protein interaction networks. PLoS Comput. Biol. 2015, 11, e1004049.
- Wang, Y.C.; Peterson, S.E.; Loring, J.F. Protein post-translational modifications and regulation of pluripotency in human stem cells. Cell Res. 2014, 24, 143–160.
- Santos, A.L.; Lindner, A.B. Protein Posttranslational Modifications: Roles in Aging and Age-Related Disease. Oxid Med. Cell Longev. 2017, 2017, 5716409.
- Zhao, S.; Xu, W.; Jiang, W.; Yu, W.; Lin, Y.; Zhang, T.; Yao, J.; Zhou, L.; Zeng, Y.; Li, H.; et al. Regulation of cellular metabolism by protein lysine acetylation. Science 2010, 327, 1000–1004.
- Liu, Y.; Tavana, O.; Gu, W. p53 modifications: Exquisite decorations of the powerful guardian. J. Mol. Cell Biol. 2019, 11, 564–577.
- Chambon, P.; Weill, J.D.; Mandel, P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem. Biophys Res. Commun. 1963, 11, 39–43.
- D’Amours, D.; Desnoyers, S.; D’Silva, I.; Poirier, G.G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999, 342 Pt 2, 249–268.
- Hottiger, M.O.; Hassa, P.O.; Luscher, B.; Schuler, H.; Koch-Nolte, F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 2010, 35, 208–219.
- Li, N.; Chen, J. ADP-ribosylation: Activation, recognition, and removal. Mol. Cells 2014, 37, 9–16.
- Gibson, B.A.; Kraus, W.L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012, 13, 411–424.
- Ummarino, S.; Hausman, C.; Di Ruscio, A. The PARP Way to Epigenetic Changes. Genes 2021, 12, 446.
- Luo, X.; Kraus, W.L. On PAR with PARP: Cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012, 26, 417–432.
- Tallis, M.; Morra, R.; Barkauskaite, E.; Ahel, I. Poly(ADP-ribosyl)ation in regulation of chromatin structure and the DNA damage response. Chromosoma 2014, 123, 79–90.
- Jungmichel, S.; Rosenthal, F.; Altmeyer, M.; Lukas, J.; Hottiger, M.O.; Nielsen, M.L. Proteome-wide identification of poly(ADP-Ribosyl)ation targets in different genotoxic stress responses. Mol. Cell 2013, 52, 272–285.
- Ke, Y.; Wang, C.; Zhang, J.; Zhong, X.; Wang, R.; Zeng, X.; Ba, X. The Role of PARPs in Inflammation-and Metabolic-Related Diseases: Molecular Mechanisms and Beyond. Cells 2019, 8, 1047.
- Chiu, L.Y.; Huang, D.Y.; Lin, W.W. PARP-1 regulates inflammasome activity by poly-ADP-ribosylation of NLRP3 and interaction with TXNIP in primary macrophages. Cell. Mol. Life Sci. 2022, 79, 108.
- Luo, X.; Ryu, K.W.; Kim, D.S.; Nandu, T.; Medina, C.J.; Gupte, R.; Gibson, B.A.; Soccio, R.E.; Yu, Y.; Gupta, R.K.; et al. PARP-1 Controls the Adipogenic Transcriptional Program by PARylating C/EBPbeta and Modulating Its Transcriptional Activity. Mol. Cell 2017, 65, 260–271.
- Wang, Y.; An, R.; Umanah, G.K.; Park, H.; Nambiar, K.; Eacker, S.M.; Kim, B.; Bao, L.; Harraz, M.M.; Chang, C.; et al. A nuclease that mediates cell death induced by DNA damage and poly(ADP-ribose) polymerase-1. Science 2016, 354, aad6872.
- Langelier, M.F.; Pascal, J.M. PARP-1 mechanism for coupling DNA damage detection to poly(ADP-ribose) synthesis. Curr. Opin. Struct. Biol. 2013, 23, 134–143.
- Kim, M.Y.; Zhang, T.; Kraus, W.L. Poly(ADP-ribosyl)ation by PARP-1: ’PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 2005, 19, 1951–1967.
- Wang, Y.; Luo, W.; Wang, Y. PARP-1 and its associated nucleases in DNA damage response. DNA Repair 2019, 81, 102651.
- Kim, M.Y.; Mauro, S.; Gevry, N.; Lis, J.T.; Kraus, W.L. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell 2004, 119, 803–814.
- Wei, H.T.; Yu, X.C. Functions of PARylation in DNA Damage Repair Pathways. Genom. Proteom. Bioinform. 2016, 14, 131–139.
- Krishnakumar, R.; Kraus, W.L. The PARP side of the nucleus: Molecular actions, physiological outcomes, and clinical targets. Mol. Cell 2010, 39, 8–24.
- Krietsch, J.; Rouleau, M.; Pic, E.; Ethier, C.; Dawson, T.M.; Dawson, V.L.; Masson, J.Y.; Poirier, G.G.; Gagne, J.P. Reprogramming cellular events by poly(ADP-ribose)-binding proteins. Mol. Aspects Med. 2013, 34, 1066–1087.
- Aberle, L.; Kruger, A.; Reber, J.M.; Lippmann, M.; Hufnagel, M.; Schmalz, M.; Trussina, I.; Schlesiger, S.; Zubel, T.; Schutz, K.; et al. PARP1 catalytic variants reveal branching and chain length-specific functions of poly(ADP-ribose) in cellular physiology and stress response. Nucleic Acids Res. 2020, 48, 10015–10033.
- Caldecott, K.W. Single-strand break repair and genetic disease. Nat. Rev. Genet. 2008, 9, 619–631.
- Koczor, C.A.; Saville, K.M.; Andrews, J.F.; Clark, J.; Fang, Q.; Li, J.; Al-Rahahleh, R.Q.; Ibrahim, M.; McClellan, S.; Makarov, M.V.; et al. Temporal dynamics of base excision/single-strand break repair protein complex assembly/disassembly are modulated by the PARP/NAD(+)/SIRT6 axis. Cell Rep. 2021, 37, 109917.
- Hoch, N.C.; Hanzlikova, H.; Rulten, S.L.; Tetreault, M.; Komulainen, E.; Ju, L.; Hornyak, P.; Zeng, Z.; Gittens, W.; Rey, S.A.; et al. XRCC1 mutation is associated with PARP1 hyperactivation and cerebellar ataxia. Nature 2017, 541, 87–91.
- Kusakabe, M.; Onishi, Y.; Tada, H.; Kurihara, F.; Kusao, K.; Furukawa, M.; Iwai, S.; Yokoi, M.; Sakai, W.; Sugasawa, K. Mechanism and regulation of DNA damage recognition in nucleotide excision repair. Genes Environ. 2019, 41, 2.
- Jeong, S.; Chung, Y.; Park, S.; Lee, S.; Choi, N.; Park, J.K. Combined treatment of ginsenoside Rg2 and piceatannol mixture reduces the apoptosis and DNA damage induced by UVB in HaCaT cells. Mol. Cell. Toxicol. 2022.
- Bergink, S.; Toussaint, W.; Luijsterburg, M.S.; Dinant, C.; Alekseev, S.; Hoeijmakers, J.H.; Dantuma, N.P.; Houtsmuller, A.B.; Vermeulen, W. Recognition of DNA damage by XPC coincides with disruption of the XPC-RAD23 complex. J. Cell Biol. 2012, 196, 681–688.
- Maltseva, E.A.; Rechkunova, N.I.; Sukhanova, M.V.; Lavrik, O.I. Poly(ADP-ribose) Polymerase 1 Modulates Interaction of the Nucleotide Excision Repair Factor XPC-RAD23B with DNA via Poly(ADP-ribosyl)ation. J. Biol. Chem. 2015, 290, 21811–21820.
- Luijsterburg, M.S.; Lindh, M.; Acs, K.; Vrouwe, M.G.; Pines, A.; van Attikum, H.; Mullenders, L.H.; Dantuma, N.P. DDB2 promotes chromatin decondensation at UV-induced DNA damage. J. Cell Biol. 2012, 197, 267–281.
- Khanna, K.K.; Jackson, S.P. DNA double-strand breaks: Signaling, repair and the cancer connection. Nat. Genet. 2001, 27, 247–254.
- Caron, M.C.; Sharma, A.K.; O’Sullivan, J.; Myler, L.R.; Ferreira, M.T.; Rodrigue, A.; Coulombe, Y.; Ethier, C.; Gagne, J.P.; Langelier, M.F.; et al. Poly(ADP-ribose) polymerase-1 antagonizes DNA resection at double-strand breaks. Nat. Commun. 2019, 10, 2954.
- Bryant, H.E.; Petermann, E.; Schultz, N.; Jemth, A.S.; Loseva, O.; Issaeva, N.; Johansson, F.; Fernandez, S.; McGlynn, P.; Helleday, T. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009, 28, 2601–2615.
- Li, M.; Yu, X. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell 2013, 23, 693–704.
- Gong, T.; Jia, B.; Gu, L.; Yu, T. KLF5-trancripted miR-125b-5p is involved in enhancing the radio-sensitivity of breast cancer cells by targeting BRCA1. Mol. Cell. Toxicol. 2022, 18, 101–110.
- Hu, Y.; Petit, S.A.; Ficarro, S.B.; Toomire, K.J.; Xie, A.; Lim, E.; Cao, S.A.; Park, E.; Eck, M.J.; Scully, R.; et al. PARP1-driven poly-ADP-ribosylation regulates BRCA1 function in homologous recombination-mediated DNA repair. Cancer Discov. 2014, 4, 1430–1447.
- Couto, C.A.; Wang, H.Y.; Green, J.C.; Kiely, R.; Siddaway, R.; Borer, C.; Pears, C.J.; Lakin, N.D. PARP regulates nonhomologous end joining through retention of Ku at double-strand breaks. J. Cell Biol. 2011, 194, 367–375.
- Poirier, G.G.; de Murcia, G.; Jongstra-Bilen, J.; Niedergang, C.; Mandel, P. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc. Natl. Acad. Sci. USA 1982, 79, 3423–3427.
- Messner, S.; Altmeyer, M.; Zhao, H.; Pozivil, A.; Roschitzki, B.; Gehrig, P.; Rutishauser, D.; Huang, D.; Caflisch, A.; Hottiger, M.O. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 2010, 38, 6350–6362.
- Aubin, R.J.; Frechette, A.; de Murcia, G.; Mandel, P.; Lord, A.; Grondin, G.; Poirier, G.G. Correlation between endogenous nucleosomal hyper(ADP-ribosyl)ation of histone H1 and the induction of chromatin relaxation. EMBO J. 1983, 2, 1685–1693.
- Ahel, D.; Horejsi, Z.; Wiechens, N.; Polo, S.E.; Garcia-Wilson, E.; Ahel, I.; Flynn, H.; Skehel, M.; West, S.C.; Jackson, S.P.; et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 2009, 325, 1240–1243.
- Chou, D.M.; Adamson, B.; Dephoure, N.E.; Tan, X.; Nottke, A.C.; Hurov, K.E.; Gygi, S.P.; Colaiacovo, M.P.; Elledge, S.J. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. USA 2010, 107, 18475–18480.
- Lee, S.G.; Kim, N.; Kim, S.M.; Park, I.B.; Kim, H.; Kim, S.; Kim, B.G.; Hwang, J.M.; Baek, I.J.; Gartner, A.; et al. Ewing sarcoma protein promotes dissociation of poly(ADP-ribose) polymerase 1 from chromatin. EMBO Rep. 2020, 21, e48676.
- Muthurajan, U.M.; Hepler, M.R.; Hieb, A.R.; Clark, N.J.; Kramer, M.; Yao, T.; Luger, K. Automodification switches PARP-1 function from chromatin architectural protein to histone chaperone. Proc. Natl. Acad. Sci. USA 2014, 111, 12752–12757.
- O’Sullivan, J.; Tedim Ferreira, M.; Gagne, J.P.; Sharma, A.K.; Hendzel, M.J.; Masson, J.Y.; Poirier, G.G. Emerging roles of eraser enzymes in the dynamic control of protein ADP-ribosylation. Nat. Commun. 2019, 10, 1182.
- Oka, S.; Kato, J.; Moss, J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 2006, 281, 705–713.
- Kawaichi, M.; Oka, J.; Zhang, J.; Ueda, K.; Hayaishi, O. Properties of poly(ADP-ribose) synthetase and ADP-ribosyl histone splitting enzyme. Princess Takamatsu. Symp. 1983, 13, 121–128.
This entry is offline, you can click here to edit this entry!
