Telomeres are essential structures formed from satellite DNA repeats at the ends of chromosomes in most eukaryotes. Satellite DNA repeat sequences are useful markers for karyotyping, but have a more enigmatic role in the eukaryotic cell. More research is needed until there is a complete picture of the biological function of telomere and other DNA satellite sequences, including chromatin structure, chromosome end-protection and species evolution with a particular focus on non-model organisms. The first problem to solve is the identification of telomere repeats, because telomere repeat identity is the foundation for any hypothesis about telomere maintenance and structure, or binding of specific proteins. Celebrating Gregor Mendel’s anniversary by going to the principles behind the experiments, a selection of recent developments and underexplored areas of research from the past are illustrated in plants and insects. Indeed, much recent work has expanded beyond the human and yeast models traditional in telomere research. Classic methods from the past, and cutting-edge in silico methods are described. These do not require specialized equipment or expensive materials and can be used, often in combination, to aid research into telomeres and satellites. This can both enrich the general understanding of chromosome maintenance mechanisms and further explore the evolution of telomeres and telomerases.
- satellite
- telomere evolution
- interstitial telomere sequences
- telomerase RNA
- NGS
- eukaryotic tree of life
1. Introduction
2. Telomeres as Steps in Species Evolution
3. Telomere Minisatellites Are Much like Any Other DNA Sequences
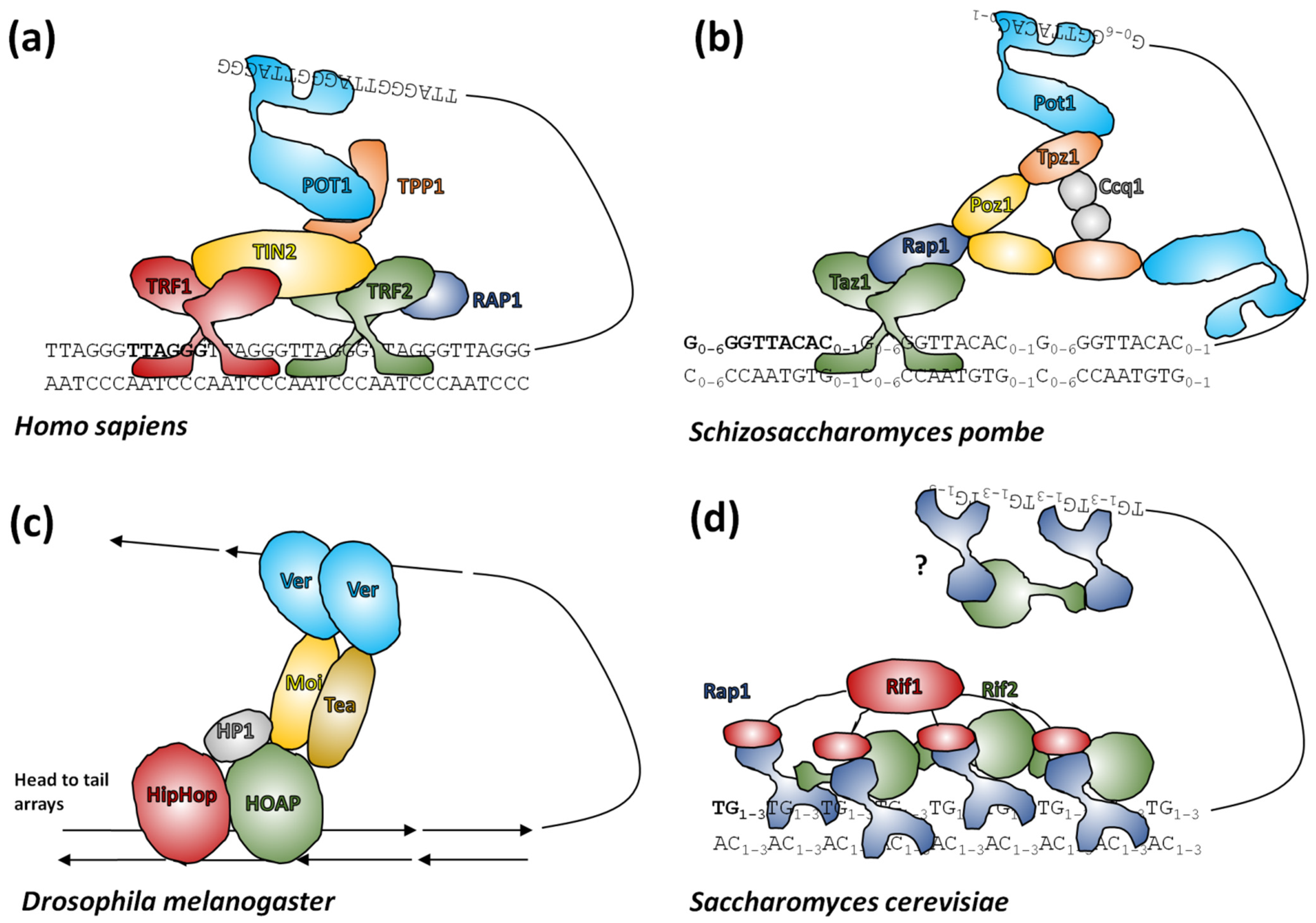
4. Telomere Proteins
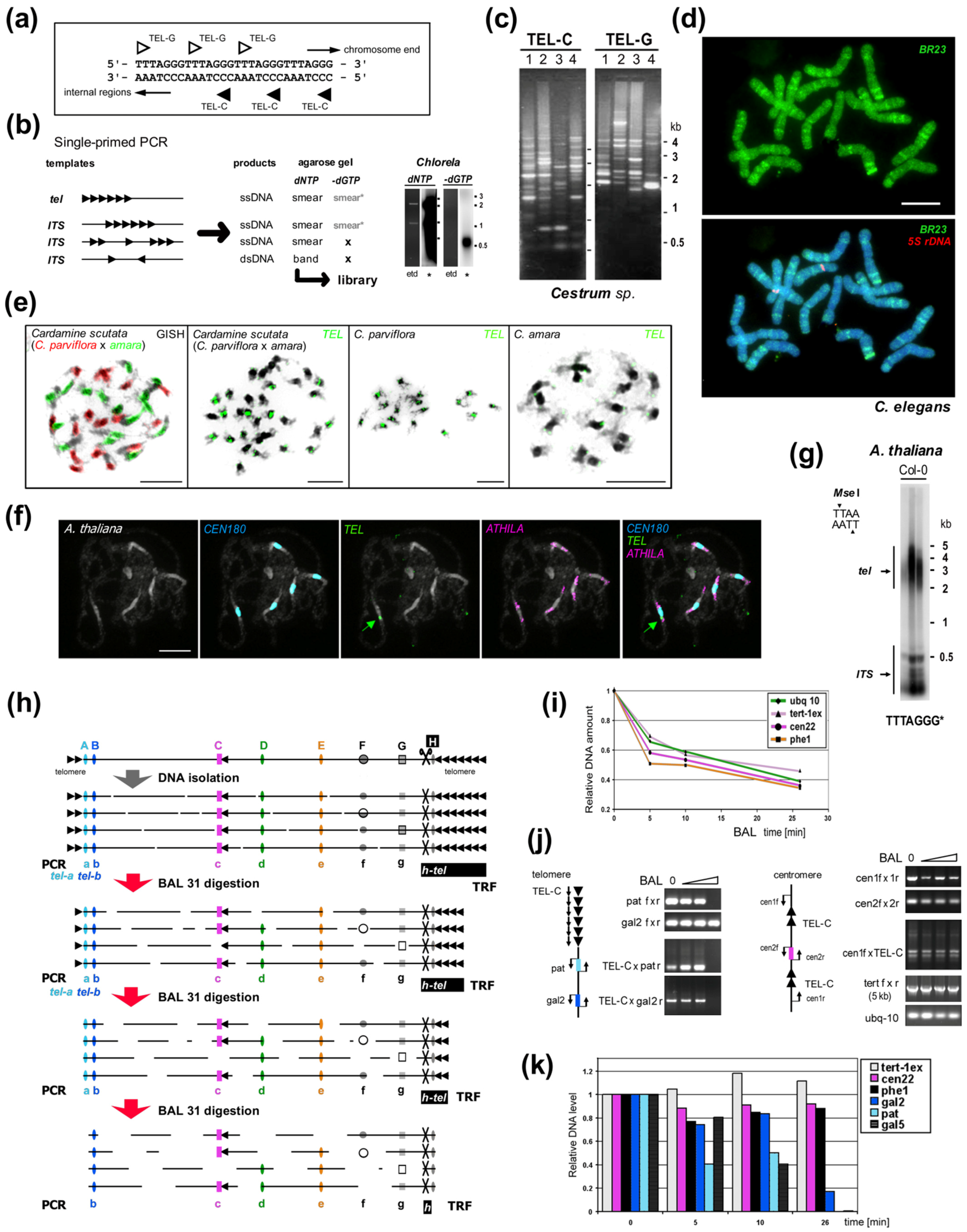
5. How to Find a Telomere Candidate
Experimental approaches which have been used successfully in the past to characterize telomeres de novo (summarized in [162]) comprise proof of the end-protection function of newly-discovered sequence in vivo [163][164], genomic DNA library screening with verification of terminal position by BAL31 digestion and Southern hybridization [16][17][62][80],[165],[166] cloning of telomerase products [54][55][58][64][167][168] and a novel combination of genomic and transcriptomic studies with classical methods [62][162][166][169]. Today raw data or assembled contigs generated by researchers or from public NGS (next generation sequencing) datasets can be mined for repetitive sequences using, e.g., Tandem Repeats Finder [170] and/or RepeatExplorer [171]. New ways of in silico analysis in combination with experimental approaches for the identification and verification of novel telomere sequences were used e.g., in yeast Lachancea sp. [47] beetle Anoplotrupes stercorosus [67], a plant with human-like telomere sequence Zostera marina (Alismatales) [172] and also a plant with unusual telomere type A. cepa [62]. Moreover, comparative transcriptome study led to identification of telomerase RNA (TR) subunits and telomeric repeats across the entire land plant phylogeny [169]. Subsequently, a new bioinformatic approach based on prediction of TR subunits in combination with results from Tandem Repeats Finder resulted in a broad identification of telomere sequences in green algae, ciliates and Stramenopiles including novel types TATAGGG, TGTTAGGG, TGTAAGGG and demonstrated the deep evolutionary TR origin in the megagroup Diaphoretickes [173].
This entry is adapted from the peer-reviewed paper 10.3390/genes13091663
References
- Kreplak, J.; Madoui, M.A.; Capal, P.; Novak, P.; Labadie, K.; Aubert, G.; Bayer, P.E.; Gali, K.K.; Syme, R.A.; Main, D.; et al. A reference genome for pea provides insight into legume genome evolution. Nat. Genet. 2019, 51, 1411–1422.
- The Honeybee Genome Sequencing Consortium. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 2006, 443, 931–949.
- McClintock, B. The fusion of broken chromosome ends of sister half-chromatids following chromatid breakage at meiotic anaphases. MO Agric. Exp. Stn. Res. Bull. 1938, 290, 1–48.
- Muller, H. The remaking of chromosomes. Collect. Net. 1938, 13, 182–198.
- Biscotti, M.A.; Olmo, E.; Heslop-Harrison, J.S. Repetitive DNA in eukaryotic genomes. Chromosome Res. 2015, 23, 415–420.
- Thakur, J.; Packiaraj, J.; Henikoff, S. Sequence, Chromatin and Evolution of Satellite DNA. Int. J. Mol. Sci. 2021, 22, 4309.
- Peska, V.; Garcia, S. Origin, Diversity, and Evolution of Telomere Sequences in Plants. Front. Plant Sci. 2020, 11, 117.
- Mikhail, F.M. Chromosomal Basis of Inheritance. In Emery and Rimoin’s Principles and Practice of Medical Genetics and Genomics, 7th ed.; Pyeritz, R.E., Korf, B.R., Grody, W.W., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 237–265.
- Tomaska, L.; Cesare, A.J.; AlTurki, T.M.; Griffith, J.D. Twenty years of t-loops: A case study for the importance of collaboration in molecular biology. DNA Repair 2020, 94, 102901.
- Bryan, T.M. G-Quadruplexes at Telomeres: Friend or Foe? Molecules 2020, 25, 3686.
- Mason, J.M.; Randall, T.A.; Capkova Frydrychova, R. Telomerase lost? Chromosoma 2016, 125, 65–73.
- Muyle, A.; Marais, G.A.B.; Bacovsky, V.; Hobza, R.; Lenormand, T. Dosage compensation evolution in plants: Theories, controversies and mechanisms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2022, 377, 20210222.
- Houben, A.; Banaei-Moghaddam, A.M.; Klemme, S.; Timmis, J.N. Evolution and biology of supernumerary B chromosomes. Cell Mol. Life Sci. 2014, 71, 467–478.
- Meyne, J.; Ratliff, R.L.; Moyzis, R.K. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc. Natl. Acad. Sci. USA 1989, 86, 7049–7053.
- Fuchs, J.; Brandes, A.; Schubert, I. Telomere sequence localization and karyotype evolution in higher plants. Plant Syst. Evol. 1995, 196, 227–241.
- Okazaki, S.; Tsuchida, K.; Maekawa, H.; Ishikawa, H.; Fujiwara, H. Identification of a pentanucleotide telomeric sequence, (TTAGG)n, in the silkworm Bombyx mori and in other insects. Mol. Cell. Biol. 1993, 13, 1424–1432.
- Richards, E.J.; Ausubel, F.M. Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell 1988, 53, 127–136.
- Sahara, K.; Marec, F.; Traut, W. TTAGG telomeric repeats in chromosomes of some insects and other arthropods. Chromosome Res. 1999, 7, 449–460.
- Fajkus, J.; Sykorova, E.; Leitch, A.R. Telomeres in evolution and evolution of telomeres. Chromosome Res. 2005, 13, 469–479.
- Blackburn, E.H.; Gall, J.G. Tandemly Repeated Sequence at Termini of Extrachromosomal Ribosomal-Rna Genes in Tetrahymena. J. Mol. Biol. 1978, 120, 33–53.
- Greider, C.W.; Blackburn, E.H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 1985, 43, 405–413.
- Greider, C.W.; Blackburn, E.H. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell 1987, 51, 887–898.
- Shay, J.W.; Wright, W.E. Telomeres and telomerase: Three decades of progress. Nat. Rev. Genet. 2019, 20, 299–309.
- Lopez, C.C.; Nielsen, L.; Edstrom, J.E. Terminal long tandem repeats in chromosomes form Chironomus pallidivittatus. Mol. Cell. Biol. 1996, 16, 3285–3290.
- Nielsen, L.; Edstrom, J.E. Complex telomere-associated repeat units in members of the genus Chironomus evolve from sequences similar to simple telomeric repeats. Mol. Cell. Biol. 1993, 13, 1583–1589.
- Roth, C.W.; Kobeski, F.; Walter, M.F.; Biessmann, H. Chromosome end elongation by recombination in the mosquito Anopheles gambiae. Mol. Cell. Biol. 1997, 17, 5176–5183.
- Madalena, C.R.; Fernandes, T.; Villasante, A.; Gorab, E. Curiously composite structures of a retrotransposon and a complex repeat associated with chromosome ends of Rhynchosciara americana (Diptera: Sciaridae). Chromosome Res. 2010, 18, 587–598.
- Rossato, R.M.; Madalena, C.R.; Gorab, E. Unusually short tandem repeats in the chromosome end structure of Rhynchosciara (Diptera: Sciaridae). Genetica 2007, 131, 109–116.
- Belfort, M.; Curcio, M.J.; Lue, N.F. Telomerase and retrotransposons: Reverse transcriptases that shaped genomes. Proc. Natl. Acad. Sci. USA 2011, 108, 20304–20310.
- Markova, D.N.; Christensen, S.M.; Betran, E. Telomere-Specialized Retroelements in Drosophila: Adaptive Symbionts of the Genome, Neutral, or in Conflict? Bioessays 2020, 42, e1900154.
- Garcia, S.; Garnatje, T.; Kovarik, A. Plant rDNA database: Ribosomal DNA loci information goes online. Chromosoma 2012, 121, 389–394.
- Sochorova, J.; Garcia, S.; Galvez, F.; Symonova, R.; Kovarik, A. Evolutionary trends in animal ribosomal DNA loci: Introduction to a new online database. Chromosoma 2018, 127, 141–150.
- Hemleben, V.; Grierson, D.; Borisjuk, N.; Volkov, R.A.; Kovarik, A. Personal Perspectives on Plant Ribosomal RNA Genes Research: From Precursor-rRNA to Molecular Evolution. Front. Plant Sci. 2021, 12, 797348.
- Aguilar, M.; Prieto, P. Telomeres and Subtelomeres Dynamics in the Context of Early Chromosome Interactions During Meiosis and Their Implications in Plant Breeding. Front. Plant Sci. 2021, 12, 672489.
- Bolzan, A.D. Chromosomal aberrations involving telomeres and interstitial telomeric sequences. Mutagenesis 2012, 27, 1–15.
- Carlton, P.M.; Cande, W.Z. Telomeres act autonomously in maize to organize the meiotic bouquet from a semipolarized chromosome orientation. J. Cell Biol. 2002, 157, 231–242.
- Cowan, C.R.; Carlton, P.M.; Cande, W.Z. The polar arrangement of telomeres in interphase and meiosis. Rabl organization and the bouquet. Plant Physiol. 2001, 125, 532–538.
- Krejci, K.; Stentoft, J.; Koch, J. Molecular cytogenetics investigation of the telomeres in a case of Philadelphia positive B-ALL with a single telomere expansion. Neoplasia 1999, 1, 492–497.
- Samassekou, O.; Li, H.; Hebert, J.; Ntwari, A.; Wang, H.; Cliche, C.G.; Bouchard, E.; Huang, S.; Yan, J. Chromosome arm-specific long telomeres: A new clonal event in primary chronic myelogenous leukemia cells. Neoplasia 2011, 13, 550–560.
- Schober, H.; Kalck, V.; Vega-Palas, M.A.; Van Houwe, G.; Sage, D.; Unser, M.; Gartenberg, M.R.; Gasser, S.M. Controlled exchange of chromosomal arms reveals principles driving telomere interactions in yeast. Genome Res. 2008, 18, 261–271.
- Slijepcevic, P. Telomeres and mechanisms of Robertsonian fusion. Chromosoma 1998, 107, 136–140.
- Shampay, J.; Szostak, J.W.; Blackburn, E.H. DNA sequences of telomeres maintained in yeast. Nature 1984, 310, 154–157.
- Greider, C.W. Telomere length regulation. Annu. Rev. Biochem. 1996, 65, 337–365.
- Matsumoto, T.; Fukui, K.; Niwa, O.; Sugawara, N.; Szostak, J.W.; Yanagida, M. Identification of healed terminal DNA fragments in linear minichromosomes of Schizosaccharomyces pombe. Mol. Cell. Biol. 1987, 7, 4424–4430.
- McEachern, M.J.; Hicks, J.B. Unusually large telomeric repeats in the yeast Candida albicans. Mol. Cell. Biol. 1993, 13, 551–560.
- McEachern, M.J.; Blackburn, E.H. A conserved sequence motif within the exceptionally diverse telomeric sequences of budding yeasts. Proc. Natl. Acad. Sci. USA 1994, 91, 3453–3457.
- Peska, V.; Fajkus, P.; Bubenik, M.; Brazda, V.; Bohalova, N.; Dvoracek, V.; Fajkus, J.; Garcia, S. Extraordinary diversity of telomeres, telomerase RNAs and their template regions in Saccharomycetaceae. Sci. Rep. 2021, 11, 12784.
- Cervenak, F.; Jurikova, K.; Devillers, H.; Kaffe, B.; Khatib, A.; Bonnell, E.; Sopkovicova, M.; Wellinger, R.J.; Nosek, J.; Tzfati, Y.; et al. Identification of telomerase RNAs in species of the Yarrowia clade provides insights into the co-evolution of telomerase, telomeric repeats and telomere-binding proteins. Sci. Rep. 2019, 9, 13365.
- Cervenak, F.; Sepsiova, R.; Nosek, J.; Tomaska, L. Step-by-Step Evolution of Telomeres: Lessons from Yeasts. Genome Biol. Evol. 2021, 13, evaa268.
- Sykorova, E.; Lim, K.Y.; Chase, M.W.; Knapp, S.; Leitch, I.J.; Leitch, A.R.; Fajkus, J. The absence of Arabidopsis-type telomeres in Cestrum and closely related genera Vestia and Sessea (Solanaceae): First evidence from eudicots. Plant J. 2003, 34, 283–291.
- Adams, S.P.; Hartman, T.P.; Lim, K.Y.; Chase, M.W.; Bennett, M.D.; Leitch, I.J.; Leitch, A.R. Loss and recovery of Arabidopsis-type telomere repeat sequences 5′-(TTTAGGG)(n)-3′ in the evolution of a major radiation of flowering plants. Proc. Biol. Sci. 2001, 268, 1541–1546.
- Adams, S.P.; Leitch, I.J.; Bennett, M.D.; Leitch, A.R. Aloe L.—A second plant family without (TTTAGGG)n telomeres. Chromosoma 2000, 109, 201–205.
- Frydrychova, R.; Marec, F. Repeated losses of TTAGG telomere repeats in evolution of beetles (Coleoptera). Genetica 2002, 115, 179–187.
- Fulneckova, J.; Sevcikova, T.; Fajkus, J.; Lukesova, A.; Lukes, M.; Vlcek, C.; Lang, B.F.; Kim, E.; Elias, M.; Sykorova, E. A broad phylogenetic survey unveils the diversity and evolution of telomeres in eukaryotes. Genome Biol. Evol. 2013, 5, 468–483.
- Sykorova, E.; Lim, K.Y.; Kunicka, Z.; Chase, M.W.; Bennett, M.D.; Fajkus, J.; Leitch, A.R. Telomere variability in the monocotyledonous plant order Asparagales. Proc. R. Soc. Lond. B Biol. Sci. 2003, 270, 1893–1904.
- Sykorova, E.; Fajkus, J.; Meznikova, M.; Lim, K.Y.; Neplechova, K.; Blattner, F.R.; Chase, M.W.; Leitch, A.R. Minisatellite telomeres occur in the family Alliaceae but are lost in Allium. Am. J. Bot. 2006, 93, 814–823.
- Bombarova, M.; Vitkova, M.; Spakulova, M.; Koubkova, B. Telomere analysis of platyhelminths and acanthocephalans by FISH and Southern hybridization. Genome 2009, 52, 897–903.
- Fulneckova, J.; Hasikova, T.; Fajkus, J.; Lukesova, A.; Elias, M.; Sykorova, E. Dynamic evolution of telomeric sequences in the green algal order Chlamydomonadales. Genome Biol. Evol. 2012, 4, 248–264.
- Mravinac, B.; Mestrovic, N.; Cavrak, V.V.; Plohl, M. TCAGG, an alternative telomeric sequence in insects. Chromosoma 2011, 120, 367–376.
- Vitkova, M.; Kral, J.; Traut, W.; Zrzavy, J.; Marec, F. The evolutionary origin of insect telomeric repeats, (TTAGG)n. Chromosome Res. 2005, 13, 145–156.
- Weiss, H.; Scherthan, H. Aloe spp.—Plants with vertebrate-like telomeric sequences. Chromosome Res. 2002, 10, 155–164.
- Fajkus, P.; Peska, V.; Sitova, Z.; Fulneckova, J.; Dvorackova, M.; Gogela, R.; Sykorova, E.; Hapala, J.; Fajkus, J. Allium telomeres unmasked: The unusual telomeric sequence (CTCGGTTATGGG)n is synthesized by telomerase. Plant J. 2016, 85, 337–347.
- Friesen, N.; Fritsch, R.M.; Blattner, F.R. Phylogeny and new intrageneric classification of Allium (Alliaceae) based on nuclear rDNA ITS sequences. Aliso 2006, 22, 372–395.
- Fulneckova, J.; Sevcikova, T.; Lukesova, A.; Sykorova, E. Transitions between the Arabidopsis-type and the human-type telomere sequence in green algae (clade Caudivolvoxa, Chlamydomonadales). Chromosoma 2016, 125, 437–451.
- Derelle, R.; Torruella, G.; Klimes, V.; Brinkmann, H.; Kim, E.; Vlcek, C.; Lang, B.F.; Elias, M. Bacterial proteins pinpoint a single eukaryotic root. Proc. Natl. Acad. Sci. USA 2015, 112, E693–E699.
- Elias, M. Protist diversity: Novel groups enrich the algal tree of life. Curr. Biol. 2021, 31, R733–R735.
- Prusakova, D.; Peska, V.; Pekar, S.; Bubenik, M.; Cizek, L.; Bezdek, A.; Capkova Frydrychova, R. Telomeric DNA sequences in beetle taxa vary with species richness. Sci. Rep. 2021, 11, 13319.
- Kuznetsova, V.; Maryańska-Nadachowska, A.; Anokhin, B.; Shapoval, N.; Shapoval, A. Chromosomal analysis of eight species of dragonflies (Anisoptera) and damselflies (Zygoptera) using conventional cytogenetics and fluorescence in situ hybridization: Insights into the karyotype evolution of the ancient insect order Odonata. J. Zool. Syst. Evol. Res. 2021, 59, 387–399.
- Gorab, E. Chromosome End Diversification in Sciarid Flies. Cells 2020, 9, 2425.
- Zhou, Y.; Wang, Y.; Xiong, X.; Appel, A.G.; Zhang, C.; Wang, X. Profiles of telomeric repeats in Insecta reveal diverse forms of telomeric motifs in Hymenopterans. Life Sci. Alliance 2022, 5.
- Aksenova, A.Y.; Mirkin, S.M. At the Beginning of the End and in the Middle of the Beginning: Structure and Maintenance of Telomeric DNA Repeats and Interstitial Telomeric Sequences. Genes 2019, 10, 118.
- Flint, J.; Bates, G.P.; Clark, K.; Dorman, A.; Willingham, D.; Roe, B.A.; Micklem, G.; Higgs, D.R.; Louis, E.J. Sequence comparison of human and yeast telomeres identifies structurally distinct subtelomeric domains. Hum. Mol. Genet. 1997, 6, 1305–1313.
- Sykorova, E.; Lim, K.Y.; Fajkus, J.; Leitch, A.R. The signature of the Cestrum genome suggests an evolutionary response to the loss of (TTTAGGG)n telomeres. Chromosoma 2003, 112, 164–172.
- Majerova, E.; Mandakova, T.; Vu, G.T.; Fajkus, J.; Lysak, M.A.; Fojtova, M. Chromatin features of plant telomeric sequences at terminal vs. internal positions. Front. Plant Sci. 2014, 5, 593.
- Mandakova, T.; Zozomova-Lihova, J.; Kudoh, H.; Zhao, Y.; Lysak, M.A.; Marhold, K. The story of promiscuous crucifers: Origin and genome evolution of an invasive species, Cardamine occulta (Brassicaceae), and its relatives. Ann. Bot. 2019, 124, 209–220.
- Naish, M.; Alonge, M.; Wlodzimierz, P.; Tock, A.J.; Abramson, B.W.; Schmucker, A.; Mandakova, T.; Jamge, B.; Lambing, C.; Kuo, P.; et al. The genetic and epigenetic landscape of the Arabidopsis centromeres. Science 2021, 374, eabi7489.
- Pich, U.; Schubert, I. Terminal heterochromatin and alternative telomeric sequences in Allium cepa. Chromosome Res. 1998, 6, 315–321.
- Sykorova, E.; Fojtova, M.; Peska, V. A polymerase chain reaction-based approach for evaluation of telomere-associated sequences and interstitial telomeric sequences. Anal. Biochem. 2013, 439, 8–10.
- Pfaffl, M.W. Quantification strategies in real-time PCR. In A–Z of Quantitative PCR; Bustin, S.A., Ed.; International University Line: La Jolla, CA, USA, 2004; pp. 87–112.
- Moyzis, R.K.; Buckingham, J.M.; Cram, L.S.; Dani, M.; Deaven, L.L.; Jones, M.D.; Meyne, J.; Ratliff, R.L.; Wu, J.R. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. USA 1988, 85, 6622–6626.
- Meyne, J.; Baker, R.J.; Hobart, H.H.; Hsu, T.C.; Ryder, O.A.; Ward, O.G.; Wiley, J.E.; Wurster-Hill, D.H.; Yates, T.L.; Moyzis, R.K. Distribution of non-telomeric sites of the (TTAGGG)n telomeric sequence in vertebrate chromosomes. Chromosoma 1990, 99, 3–10.
- Maravilla, A.J.; Rosato, M.; Alvarez, I.; Nieto Feliner, G.; Rossello, J.A. Interstitial Arabidopsis-Type Telomeric Repeats in Asteraceae. Plants 2021, 10, 2794.
- Richards, E.J.; Goodman, H.M.; Ausubel, F.M. The centromere region of Arabidopsis thaliana chromosome 1 contains telomere-similar sequences. Nucleic Acids Res. 1991, 19, 3351–3357.
- Luo, X.; He, Z.; Liu, J.; Wu, H.; Gong, X. FISH Mapping of Telomeric and Non-Telomeric (AG3T3)3 Reveal the Chromosome Numbers and Chromosome Rearrangements of 41 Woody Plants. Genes 2022, 13, 1239.
- Rovatsos, M.; Kratochvil, L.; Altmanova, M.; Johnson Pokorna, M. Interstitial Telomeric Motifs in Squamate Reptiles: When the Exceptions Outnumber the Rule. PLoS ONE 2015, 10, e0134985.
- Rego, A.; Marec, F. Telomeric and interstitial telomeric sequences in holokinetic chromosomes of Lepidoptera: Telomeric DNA mediates association between postpachytene bivalents in achiasmatic meiosis of females. Chromosome Res. 2003, 11, 681–694.
- Azzalin, C.M.; Nergadze, S.G.; Giulotto, E. Human intrachromosomal telomeric-like repeats: Sequence organization and mechanisms of origin. Chromosoma 2001, 110, 75–82.
- Chirino, M.G.; Dalikova, M.; Marec, F.R.; Bressa, M.J. Chromosomal distribution of interstitial telomeric sequences as signs of evolution through chromosome fusion in six species of the giant water bugs (Hemiptera, Belostoma). Ecol. Evol. 2017, 7, 5227–5235.
- Gaspin, C.; Rami, J.F.; Lescure, B. Distribution of short interstitial telomere motifs in two plant genomes: Putative origin and function. BMC Plant Biol. 2010, 10, 283.
- Aksenova, A.Y.; Han, G.; Shishkin, A.A.; Volkov, K.V.; Mirkin, S.M. Expansion of Interstitial Telomeric Sequences in Yeast. Cell Rep. 2015, 13, 1545–1551.
- Nergadze, S.G.; Rocchi, M.; Azzalin, C.M.; Mondello, C.; Giulotto, E. Insertion of telomeric repeats at intrachromosomal break sites during primate evolution. Genome Res. 2004, 14, 1704–1710.
- Ijdo, J.W.; Baldini, A.; Ward, D.C.; Reeders, S.T.; Wells, R.A. Origin of human chromosome 2: An ancestral telomere-telomere fusion. Proc. Natl. Acad. Sci. USA 1991, 88, 9051–9055.
- McClintock, B. The stability of broken ends of chromosomes in Zea mays. Genetics 1940, 26, 234–282.
- Bailey, S.M.; Murnane, J.P. Telomeres, chromosome instability and cancer. Nucleic Acids Res. 2006, 34, 2408–2417.
- Lowden, M.R.; Flibotte, S.; Moerman, D.G.; Ahmed, S. DNA synthesis generates terminal duplications that seal end-to-end chromosome fusions. Science 2011, 332, 468–471.
- Bosco, G.; Haber, J.E. Chromosome break-induced DNA replication leads to nonreciprocal translocations and telomere capture. Genetics 1998, 150, 1037–1047.
- Kramara, J.; Osia, B.; Malkova, A. Break-Induced Replication: The Where, The Why, and The How. Trends Genet. 2018, 34, 518–531.
- Hackett, J.A.; Feldser, D.M.; Greider, C.W. Telomere dysfunction increases mutation rate and genomic instability. Cell 2001, 106, 275–286.
- Fojtova, M.; Fulneckova, J.; Fajkus, J.; Kovarik, A. Recovery of tobacco cells from cadmium stress is accompanied by DNA repair and increased telomerase activity. J. Exp. Bot. 2002, 53, 2151–2158.
- Jankowska, M.; Fuchs, J.; Klocke, E.; Fojtova, M.; Polanska, P.; Fajkus, J.; Schubert, V.; Houben, A. Holokinetic centromeres and efficient telomere healing enable rapid karyotype evolution. Chromosoma 2015, 124, 519–528.
- Tsujimoto, H.; Yamada, T.; Sasakuma, T. Molecular structure of a wheat chromosome end healed after gametocidal gene-induced breakage. Proc. Natl. Acad. Sci. USA 1997, 94, 3140–3144.
- Farr, C.; Fantes, J.; Goodfellow, P.; Cooke, H. Functional reintroduction of human telomeres into mammalian cells. Proc. Natl. Acad. Sci. USA 1991, 88, 7006–7010.
- Wilkie, A.O.; Lamb, J.; Harris, P.C.; Finney, R.D.; Higgs, D.R. A truncated human chromosome 16 associated with alpha thalassaemia is stabilized by addition of telomeric repeat (TTAGGG)n. Nature 1990, 346, 868–871.
- Yu, W.; Lamb, J.C.; Han, F.; Birchler, J.A. Telomere-mediated chromosomal truncation in maize. Proc. Natl. Acad. Sci. USA 2006, 103, 17331–17336.
- Kapusi, E.; Ma, L.; Teo, C.H.; Hensel, G.; Himmelbach, A.; Schubert, I.; Mette, M.F.; Kumlehn, J.; Houben, A. Telomere-mediated truncation of barley chromosomes. Chromosoma 2012, 121, 181–190.
- Tamar, S.; Papadopoulou, B. A telomere-mediated chromosome fragmentation approach to assess mitotic stability and ploidy alterations of Leishmania chromosomes. J. Biol. Chem. 2001, 276, 11662–11673.
- Teo, C.H.; Ma, L.; Kapusi, E.; Hensel, G.; Kumlehn, J.; Schubert, I.; Houben, A.; Mette, M.F. Induction of telomere-mediated chromosomal truncation and stability of truncated chromosomes in Arabidopsis thaliana. Plant J. 2011, 68, 28–39.
- Tada, S.; Ohkuchi, H.; Matsushita-Morita, M.; Furukawa, I.; Hattori, R.; Suzuki, S.; Kashiwagi, Y.; Kusumoto, K. Telomere-mediated chromosomal truncation in Aspergillus oryzae. J. Biosci. Bioeng. 2015, 119, 43–46.
- Kornberg, R.D. Structure of chromatin. Annu. Rev. Biochem. 1977, 46, 931–954.
- Fajkus, J.; Trifonov, E.N. Columnar packing of telomeric nucleosomes. Biochem. Biophys. Res. Commun. 2001, 280, 961–963.
- Soman, A.; Wong, S.Y.; Korolev, N.; Surya, W.; Lattman, S.; Vogirala, V.K.; Chen, Q.; Berezhnoy, N.V.; van Noort, J.; Rhodes, D.; et al. The columnar structure of human telomeric chromatin suggests mechanisms for telomere maintenance. bioRxiv 2022.
- De Lange, T. Shelterin-Mediated Telomere Protection. Annu. Rev. Genet. 2018, 52, 223–247.
- Riha, K.; McKnight, T.D.; Fajkus, J.; Vyskot, B.; Shippen, D.E. Analysis of the G-overhang structures on plant telomeres: Evidence for two distinct telomere architectures. Plant J. 2000, 23, 633–641.
- Eberhard, S.; Valuchova, S.; Ravat, J.; Fulnecek, J.; Jolivet, P.; Bujaldon, S.; Lemaire, S.D.; Wollman, F.A.; Teixeira, M.T.; Riha, K.; et al. Molecular characterization of Chlamydomonas reinhardtii telomeres and telomerase mutants. Life Sci. Alliance 2019, 2, e201900315.
- Kazda, A.; Zellinger, B.; Rossler, M.; Derboven, E.; Kusenda, B.; Riha, K. Chromosome end protection by blunt-ended telomeres. Genes Dev. 2012, 26, 1703–1713.
- Miyake, Y.; Nakamura, M.; Nabetani, A.; Shimamura, S.; Tamura, M.; Yonehara, S.; Saito, M.; Ishikawa, F. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol. Cell 2009, 36, 193–206.
- Wellinger, R.J. The CST complex and telomere maintenance: The exception becomes the rule. Mol. Cell 2009, 36, 168–169.
- He, Y.; Song, H.; Chan, H.; Liu, B.; Wang, Y.; Susac, L.; Zhou, Z.H.; Feigon, J. Structure of Tetrahymena telomerase-bound CST with polymerase alpha-primase. Nature 2022, 608, 813–818.
- Zaug, A.J.; Goodrich, K.J.; Song, J.J.; Sullivan, A.E.; Cech, T.R. Reconstitution of a telomeric replicon organized by CST. Nature 2022, 608, 819–825.
- Rice, C.; Skordalakes, E. Structure and function of the telomeric CST complex. Comput. Struct. Biotechnol. J. 2016, 14, 161–167.
- Linger, B.R.; Price, C.M. Conservation of telomere protein complexes: Shuffling through evolution. Crit. Rev. Biochem. Mol. Biol. 2009, 44, 434–446.
- Chandra, A.; Hughes, T.R.; Nugent, C.I.; Lundblad, V. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 2001, 15, 404–414.
- Pennock, E.; Buckley, K.; Lundblad, V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 2001, 104, 387–396.
- Wan, B.; Tang, T.; Upton, H.; Shuai, J.; Zhou, Y.; Li, S.; Chen, J.; Brunzelle, J.S.; Zeng, Z.; Collins, K.; et al. The Tetrahymena telomerase p75-p45-p19 subcomplex is a unique CST complex. Nat. Struct. Mol. Biol. 2015, 22, 1023–1026.
- Fell, V.L.; Schild-Poulter, C. The Ku heterodimer: Function in DNA repair and beyond. Mutat. Res. Rev. Mutat. Res. 2015, 763, 15–29.
- Sui, J.; Zhang, S.; Chen, B.P.C. DNA-dependent protein kinase in telomere maintenance and protection. Cell Mol. Biol. Lett. 2020, 25, 2.
- Chen, Y. The structural biology of the shelterin complex. Biol. Chem. 2019, 400, 457–466.
- Cacchione, S.; Cenci, G.; Raffa, G.D. Silence at the End: How Drosophila Regulates Expression and Transposition of Telomeric Retroelements. J. Mol. Biol. 2020, 432, 4305–4321.
- Shi, T.; Bunker, R.D.; Mattarocci, S.; Ribeyre, C.; Faty, M.; Gut, H.; Scrima, A.; Rass, U.; Rubin, S.M.; Shore, D.; et al. Rif1 and Rif2 shape telomere function and architecture through multivalent Rap1 interactions. Cell 2013, 153, 1340–1353.
- De Lange, T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 2005, 19, 2100–2110.
- Bilaud, T.; Koering, C.E.; Binet-Brasselet, E.; Ancelin, K.; Pollice, A.; Gasser, S.M.; Gilson, E. The telobox, a Myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res. 1996, 24, 1294–1303.
- Stansel, R.M.; de Lange, T.; Griffith, J.D. T-loop assembly in vitro involves binding of TRF2 near the 3’ telomeric overhang. EMBO J. 2001, 20, 5532–5540.
- Erdel, F.; Kratz, K.; Willcox, S.; Griffith, J.D.; Greene, E.C.; de Lange, T. Telomere Recognition and Assembly Mechanism of Mammalian Shelterin. Cell Rep. 2017, 18, 41–53.
- Zaug, A.J.; Podell, E.R.; Nandakumar, J.; Cech, T.R. Functional interaction between telomere protein TPP1 and telomerase. Genes Dev. 2010, 24, 613–622.
- Raffa, G.D.; Raimondo, D.; Sorino, C.; Cugusi, S.; Cenci, G.; Cacchione, S.; Gatti, M.; Ciapponi, L. Verrocchio, a Drosophila OB fold-containing protein, is a component of the terminin telomere-capping complex. Genes Dev. 2010, 24, 1596–1601.
- Miyoshi, T.; Kanoh, J.; Saito, M.; Ishikawa, F. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science 2008, 320, 1341–1344.
- Xue, J.; Chen, H.; Wu, J.; Takeuchi, M.; Inoue, H.; Liu, Y.; Sun, H.; Chen, Y.; Kanoh, J.; Lei, M. Structure of the fission yeast S. pombe telomeric Tpz1-Poz1-Rap1 complex. Cell Res. 2017, 27, 1503–1520.
- Shi, K.; Huang, W.M.; Aihara, H. An enzyme-catalyzed multistep DNA refolding mechanism in hairpin telomere formation. PLoS Biol. 2013, 11, e1001472.
- Wellinger, R.J.; Zakian, V.A. Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: Beginning to end. Genetics 2012, 191, 1073–1105.
- Prochazkova Schrumpfova, P.; Fojtova, M.; Fajkus, J. Telomeres in Plants and Humans: Not So Different, Not So Similar. Cells 2019, 8, 58.
- Peska, V.; Schrumpfova, P.P.; Fajkus, J. Using the telobox to search for plant telomere binding proteins. Curr. Protein Pept. Sci. 2011, 12, 75–83.
- Prochazkova Schrumpfova, P.; Schorova, S.; Fajkus, J. Telomere- and Telomerase-Associated Proteins and Their Functions in the Plant Cell. Front. Plant Sci. 2016, 7, 851.
- Fulcher, N.; Riha, K. Using Centromere Mediated Genome Elimination to Elucidate the Functional Redundancy of Candidate Telomere Binding Proteins in Arabidopsis thaliana. Front. Genet. 2015, 6, 349.
- Schrumpfova, P.; Kuchar, M.; Mikova, G.; Skrisovska, L.; Kubicarova, T.; Fajkus, J. Characterization of two Arabidopsis thaliana myb-like proteins showing affinity to telomeric DNA sequence. Genome 2004, 47, 316–324.
- Mozgova, I.; Schrumpfova, P.P.; Hofr, C.; Fajkus, J. Functional characterization of domains in AtTRB1, a putative telomere-binding protein in Arabidopsis thaliana. Phytochemistry 2008, 69, 1814–1819.
- Schrumpfova, P.P.; Vychodilova, I.; Dvorackova, M.; Majerska, J.; Dokladal, L.; Schorova, S.; Fajkus, J. Telomere repeat binding proteins are functional components of Arabidopsis telomeres and interact with telomerase. Plant J. 2014, 77, 770–781.
- Dvorackova, M.; Rossignol, P.; Shaw, P.J.; Koroleva, O.A.; Doonan, J.H.; Fajkus, J. AtTRB1, a telomeric DNA-binding protein from Arabidopsis, is concentrated in the nucleolus and shows highly dynamic association with chromatin. Plant J. 2010, 61, 637–649.
- Dreissig, S.; Schiml, S.; Schindele, P.; Weiss, O.; Rutten, T.; Schubert, V.; Gladilin, E.; Mette, M.F.; Puchta, H.; Houben, A. Live-cell CRISPR imaging in plants reveals dynamic telomere movements. Plant J. 2017, 91, 565–573.
- Khosravi, S.; Dreissig, S.; Schindele, P.; Wolter, F.; Rutten, T.; Puchta, H.; Houben, A. Live-Cell CRISPR Imaging in Plant Cells with a Telomere-Specific Guide RNA. Methods Mol. Biol. 2020, 2166, 343–356.
- Schrumpfova, P.P.; Vychodilova, I.; Hapala, J.; Schorova, S.; Dvoracek, V.; Fajkus, J. Telomere binding protein TRB1 is associated with promoters of translation machinery genes in vivo. Plant Mol. Biol. 2016, 90, 189–206.
- Zhou, Y.; Hartwig, B.; James, G.V.; Schneeberger, K.; Turck, F. Complementary Activities of TELOMERE REPEAT BINDING Proteins and Polycomb Group Complexes in Transcriptional Regulation of Target Genes. Plant Cell 2016, 28, 87–101.
- Zhou, Y.; Wang, Y.; Krause, K.; Yang, T.; Dongus, J.A.; Zhang, Y.; Turck, F. Telobox motifs recruit CLF/SWN-PRC2 for H3K27me3 deposition via TRB factors in Arabidopsis. Nat. Genet. 2018, 50, 638–644.
- Valuchova, S.; Fulnecek, J.; Prokop, Z.; Stolt-Bergner, P.; Janouskova, E.; Hofr, C.; Riha, K. Protection of Arabidopsis Blunt-Ended Telomeres Is Mediated by a Physical Association with the Ku Heterodimer. Plant Cell 2017, 29, 1533–1545.
- Hass, E.P.; Zappulla, D.C. The Ku subunit of telomerase binds Sir4 to recruit telomerase to lengthen telomeres in S. cerevisiae. eLife 2015, 4, e07750.
- Hsu, H.L.; Gilley, D.; Blackburn, E.H.; Chen, D.J. Ku is associated with the telomere in mammals. Proc. Natl. Acad. Sci. USA 1999, 96, 12454–12458.
- Hsu, H.L.; Gilley, D.; Galande, S.A.; Hande, M.P.; Allen, B.; Kim, S.H.; Li, G.C.; Campisi, J.; Kohwi-Shigematsu, T.; Chen, D.J. Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev. 2000, 14, 2807–2812.
- D’Adda di Fagagna, F.; Hande, M.P.; Tong, W.M.; Roth, D.; Lansdorp, P.M.; Wang, Z.Q.; Jackson, S.P. Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr. Biol. 2001, 11, 1192–1196.
- Samper, E.; Goytisolo, F.A.; Slijepcevic, P.; van Buul, P.P.; Blasco, M.A. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 2000, 1, 244–252.
- Fisher, T.S.; Taggart, A.K.; Zakian, V.A. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat. Struct. Mol. Biol. 2004, 11, 1198–1205.
- Larcher, M.V.; Pasquier, E.; MacDonald, R.S.; Wellinger, R.J. Ku Binding on Telomeres Occurs at Sites Distal from the Physical Chromosome Ends. PLoS Genet. 2016, 12, e1006479.
- Nelson, A.D.; Shippen, D.E. Blunt-ended telomeres: An alternative ending to the replication and end protection stories. Genes Dev. 2012, 26, 1648–1652.
- Peska, V.; Sitova, Z.; Fajkus, P.; Fajkus, J. BAL31-NGS approach for identification of telomeres de novo in large genomes. Methods 2017, 114, 16–27.
- Szostak, J.W.; Blackburn, E.H. Cloning yeast telomeres on linear plasmid vectors. Cell 1982, 29, 245–255.
- Perrot, M.; Barreau, C.; Begueret, J. Nonintegrative transformation in the filamentous fungus Podospora anserina: Stabilization of a linear vector by the chromosomal ends of Tetrahymena thermophila. Mol. Cell. Biol. 1987, 7, 1725–1730.
- Petracek, M.E.; Lefebvre, P.A.; Silflow, C.D.; Berman, J. Chlamydomonas telomere sequences are A+T-rich but contain three consecutive G-C base pairs. Proc. Natl. Acad. Sci. USA 1990, 87, 8222–8226.
- Peska, V.; Fajkus, P.; Fojtova, M.; Dvorackova, M.; Hapala, J.; Dvoracek, V.; Polanska, P.; Leitch, A.R.; Sykorova, E.; Fajkus, J. Characterisation of an unusual telomere motif (TTTTTTAGGG)n in the plant Cestrum elegans (Solanaceae), a species with a large genome. Plant J. 2015, 82, 644–654.
- Schumpert, C.; Nelson, J.; Kim, E.; Dudycha, J.L.; Patel, R.C. Telomerase activity and telomere length in Daphnia. PLoS ONE 2015, 10, e0127196.
- Uzlikova, M.; Fulneckova, J.; Weisz, F.; Sykorova, E.; Nohynkova, E.; Tumova, P. Characterization of telomeres and telomerase from the single-celled eukaryote Giardia intestinalis. Mol. Biochem. Parasitol. 2017, 211, 31–38.
- Fajkus, P.; Peska, V.; Zavodnik, M.; Fojtova, M.; Fulneckova, J.; Dobias, S.; Kilar, A.; Dvorackova, M.; Zachova, D.; Necasova, I.; et al. Telomerase RNAs in land plants. Nucleic Acids Res. 2019, 47, 9842–9856.
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580.
- Novak, P.; Neumann, P.; Macas, J. Global analysis of repetitive DNA from unassembled sequence reads using RepeatExplorer2. Nat. Protoc. 2020, 15, 3745–3776.
- Peska, V.; Matl, M.; Mandakova, T.; Vitales, D.; Fajkus, P.; Fajkus, J.; Garcia, S. Human-like telomeres in Zostera marina reveal a mode of transition from the plant to the human telomeric sequences. J. Exp. Bot. 2020, 71, 5786–5793.
- Fajkus, P.; Kilar, A.; Nelson, A.D.L.; Hola, M.; Peska, V.; Goffova, I.; Fojtova, M.; Zachova, D.; Fulneckova, J.; Fajkus, J. Evolution of plant telomerase RNAs: Farther to the past, deeper to the roots. Nucleic Acids Res. 2021, 49, 7680–7694.
