Circadian clocks evolved to enable organisms to anticipate and prepare for periodic environmental changes driven by the day–night cycle. This internal timekeeping mechanism is built on autoregulatory transcription–translation feedback loops that control the rhythmic expression of core clock genes and their protein products. The levels of clock proteins rise and ebb throughout a 24-h period through their rhythmic synthesis and destruction. In the ubiquitin–proteasome system, the process of polyubiquitination, or the covalent attachment of a ubiquitin chain, marks a protein for degradation by the 26S proteasome. The process is regulated by E3 ubiquitin ligases, which recognize specific substrates for ubiquitination.
- circadian rhythms
- E3 ubiquitin ligases
- ubiquitin proteasome system
1. Introduction
2. Mammalian Circadian Rhythms and Ubiquitin Ligases
2.1. Overview of Circadian Rhythms in Mammals
2.1.1. The Suprachiasmatic Nucleus
2.1.2. The Mammalian Core Clock Machinery
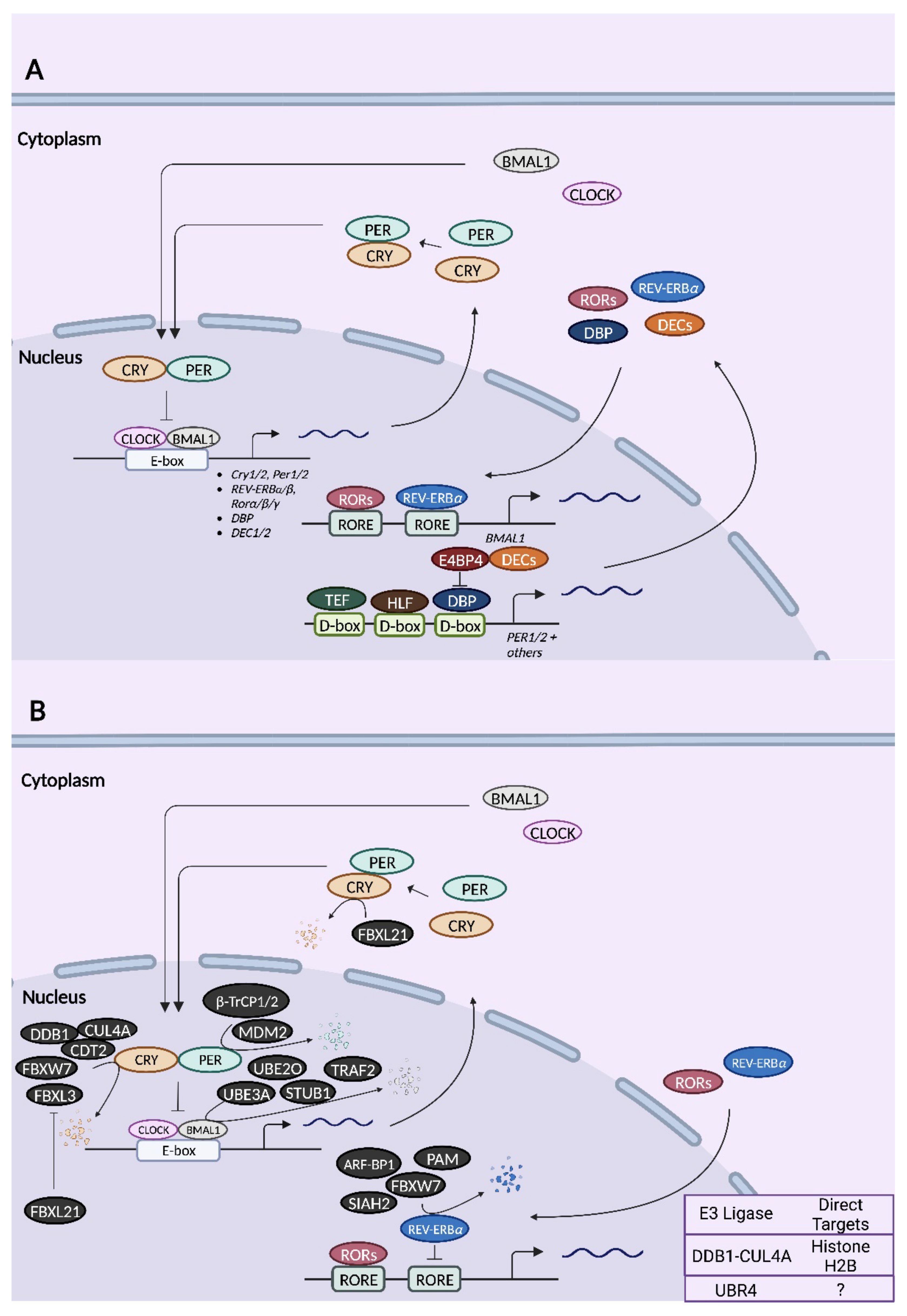
2.2. Mammalian Ubiquitin Ligases
2.2.1. β-TrCP1 (FBXW1) and β-TrCP2 (FBXW11)
2.2.2. Mouse Double Minute 2 Homolog (MDM2)
2.2.3. FBXL3
2.2.4. FBXL21
2.2.5. DDB1–CUL4A and DDB1–CUL4A–CDT2
2.2.6. FBXW7
2.2.7. TNF Receptor-Associated Factor 2 (TRAF2)
2.2.8. STIP1 Homology and U-Box-Containing Protein 1 (STUB1)
2.2.9. UBE3A
2.2.10. UBE2O
2.2.11. Seven in Absentia 2 (SIAH2)
2.2.12. ARF-BP1 and PAM (Myc-BP2)
This entry is adapted from the peer-reviewed paper 10.3390/ijms231810569
References
- Moore-Ede, M.C. Physiology of the Circadian Timing System: Predictive versus Reactive Homeostasis. Am. J. Physiol. 1986, 250, R737–R752.
- Lowrey, P.L.; Takahashi, J.S. Chapter 6—Genetics of Circadian Rhythms in Mammalian Model Organisms. In The Genetics of Circadian Rhythms; Brody, S., Ed.; Advances in Genetics; Academic Press: Cambridge, MA, USA, 2011; Volume 74, pp. 175–230.
- Buijs, F.N.; León-Mercado, L.; Guzmán-Ruiz, M.; Guerrero-Vargas, N.N.; Romo-Nava, F.; Buijs, R.M. The Circadian System: A Regulatory Feedback Network of Periphery and Brain. Physiology 2016, 31, 170–181.
- Roenneberg, T.; Merrow, M. The Circadian Clock and Human Health. Curr. Biol. 2016, 26, R432–R443.
- Ralph, M.R.; Foster, R.G.; Davis, F.C.; Menaker, M. Transplanted Suprachiasmatic Nucleus Determines Circadian Period. Science 1990, 247, 975–978.
- Golombek, D.A.; Rosenstein, R.E. Physiology of Circadian Entrainment. Physiol. Rev. 2010, 90, 1063–1102.
- Mendoza-Viveros, L.; Bouchard-Cannon, P.; Hegazi, S.; Cheng, A.H.; Pastore, S.; Cheng, H.-Y.M. Molecular Modulators of the Circadian Clock: Lessons from Flies and Mice. Cell. Mol. Life Sci. 2017, 74, 1035–1059.
- McClellan, A.J.; Laugesen, S.H.; Ellgaard, L. Cellular Functions and Molecular Mechanisms of Non-Lysine Ubiquitination. Open Biol. 2019, 9, 190147.
- Yau, R.; Rape, M. The Increasing Complexity of the Ubiquitin Code. Nat. Cell Biol. 2016, 18, 579–586.
- Kwon, Y.T.; Ciechanover, A. The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends Biochem. Sci. 2017, 42, 873–886.
- Baumeister, W.; Walz, J.; Zühl, F.; Seemüller, E. The Proteasome: Paradigm of a Self-Compartmentalizing Protease. Cell 1998, 92, 367–380.
- Liao, Y.; Sumara, I.; Pangou, E. Non-Proteolytic Ubiquitylation in Cellular Signaling and Human Disease. Commun. Biol. 2022, 5, 114.
- Kleiger, G.; Mayor, T. Perilous Journey: A Tour of the Ubiquitin-Proteasome System. Trends Cell Biol. 2014, 24, 352–359.
- Zhao, Y.; Sun, Y. Cullin-RING Ligases as Attractive Anti-Cancer Targets. Curr. Pharm. Des. 2013, 19, 3215–3225.
- Yang, Q.; Zhao, J.; Chen, D.; Wang, Y. E3 Ubiquitin Ligases: Styles, Structures and Functions. Mol. Biomed. 2021, 2, 23.
- Tasaki, T.; Sriram, S.M.; Park, K.S.; Kwon, Y.T. The N-End Rule Pathway. Annu. Rev. Biochem. 2012, 81, 261–289.
- Yang, Y.; Duguay, D.; Bédard, N.; Rachalski, A.; Baquiran, G.; Na, C.H.; Fahrenkrug, J.; Storch, K.-F.; Peng, J.; Wing, S.S.; et al. Regulation of Behavioral Circadian Rhythms and Clock Protein PER1 by the Deubiquitinating Enzyme USP2. Biol. Open 2012, 1, 789–801.
- Yang, Y.; Duguay, D.; Fahrenkrug, J.; Cermakian, N.; Wing, S.S. USP2 Regulates the Intracellular Localization of PER1 and Circadian Gene Expression. J. Biol. Rhythms 2014, 29, 243–256.
- Eletr, Z.M.; Wilkinson, K.D. Regulation of Proteolysis by Human Deubiquitinating Enzymes. Biochim. Biophys. Acta-Mol. Cell Res. 2014, 1843, 114–128.
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242.
- Sehnal, D.; Bittrich, S.; Deshpande, M.; Svobodová, R.; Berka, K.; Bazgier, V.; Velankar, S.; Burley, S.K.; Koča, J.; Rose, A.S. Mol* Viewer: Modern Web App for 3D Visualization and Analysis of Large Biomolecular Structures. Nucleic Acids Res. 2021, 49, W431–W437.
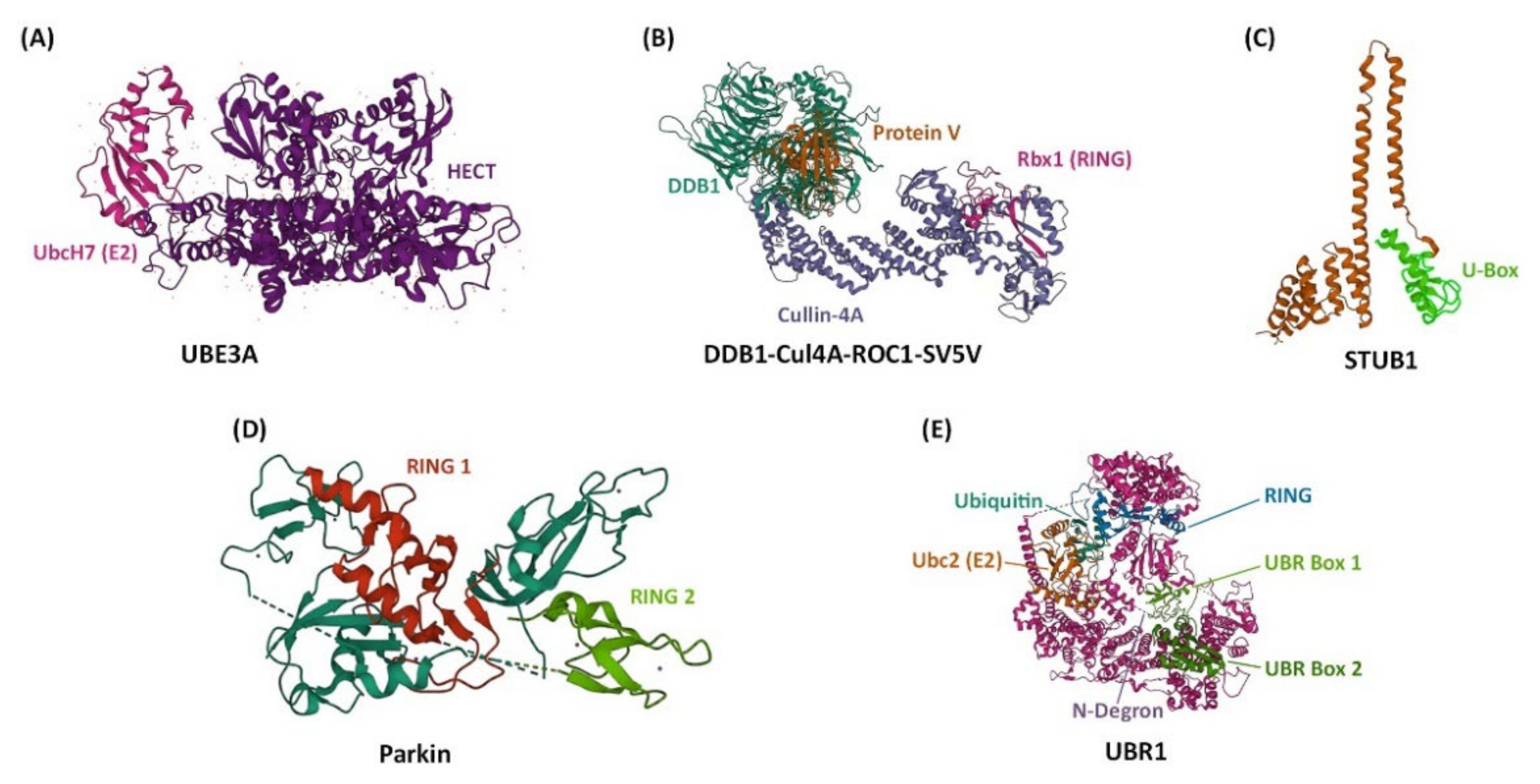
- Huang, L.; Kinnucan, E.; Wang, G.; Beaudenon, S.; Howley, P.M.; Huibregtse, J.M.; Pavletich, N.P. Structure of an E6AP-UbcH7 Complex: Insights into Ubiquitination by the E2–E3 Enzyme Cascade. Science 1999, 286, 1321–1326.
- Angers, S.; Li, T.; Yi, X.; MacCoss, M.J.; Moon, R.T.; Zheng, N. Molecular Architecture and Assembly of the DDB1–CUL4A Ubiquitin Ligase Machinery. Nature 2006, 443, 590–593.
- Zhang, M.; Windheim, M.; Roe, S.M.; Peggie, M.; Cohen, P.; Prodromou, C.; Pearl, L.H. Chaperoned Ubiquitylation—Crystal Structures of the CHIP U Box E3 Ubiquitin Ligase and a CHIP-Ubc13-Uev1a Complex. Mol. Cell 2005, 20, 525–538.
- Trempe, J.-F.; Sauvé, V.; Grenier, K.; Seirafi, M.; Tang, M.Y.; Ménade, M.; Al-Abdul-Wahid, S.; Krett, J.; Wong, K.; Kozlov, G.; et al. Structure of Parkin Reveals Mechanisms for Ubiquitin Ligase Activation. Science 2013, 340, 1451–1455.
- Pan, M.; Zheng, Q.; Wang, T.; Liang, L.; Mao, J.; Zuo, C.; Ding, R.; Ai, H.; Xie, Y.; Si, D.; et al. Structural Insights into Ubr1-Mediated N-Degron Polyubiquitination. Nature 2021, 600, 334–338.
- Abrahamson, E.E.; Moore, R.Y. Suprachiasmatic Nucleus in the Mouse: Retinal Innervation, Intrinsic Organization and Efferent Projections. Brain Res. 2001, 916, 172–191.
- Herzog, E.D.; Takahashi, J.S.; Block, G.D. Clock Controls Circadian Period in Isolated Suprachiasmatic Nucleus Neurons. Nat. Neurosci. 1998, 1, 708–713.
- Welsh, D.K.; Logothetis, D.E.; Meister, M.; Reppert, S.M. Individual Neurons Dissociated from Rat Suprachiasmatic Nucleus Express Independently Phased Circadian Firing Rhythms. Neuron 1995, 14, 697–706.
- Yamaguchi, S.; Isejima, H.; Matsuo, T.; Okura, R.; Yagita, K.; Kobayashi, M.; Okamura, H. Synchronization of Cellular Clocks in the Suprachiasmatic Nucleus. Science 2003, 302, 1408–1412.
- Maywood, E.S.; Chesham, J.E.; Brien, J.A.; Hastings, M.H. A Diversity of Paracrine Signals Sustains Molecular Circadian Cycling in Suprachiasmatic Nucleus Circuits. Proc. Natl. Acad. Sci. USA 2011, 108, 14306–14311.
- Varadarajan, S.; Tajiri, M.; Jain, R.; Holt, R.; Ahmed, Q.; LeSauter, J.; Silver, R. Connectome of the Suprachiasmatic Nucleus: New Evidence of the Core-Shell Relationship. eNeuro 2018, 5, ENEURO.0205-18.2018.
- Ono, D.; Honma, K.; Yanagawa, Y.; Yamanaka, A.; Honma, S. GABA in the Suprachiasmatic Nucleus Refines Circadian Output Rhythms in Mice. Commun. Biol. 2019, 2, 232.
- Ashton, A.; Foster, R.G.; Jagannath, A. Photic Entrainment of the Circadian System. Int. J. Mol. Sci. 2022, 23, 729.
- Hughes, S.; Jagannath, A.; Hankins, M.W.; Foster, R.G.; Peirson, S.N. Chapter Six—Photic Regulation of Clock Systems. In Methods in Enzymology; Sehgal, A., Ed.; Circadian Rhythms and Biological Clocks, Part B; Academic Press: Cambridge, MA, USA, 2015; Volume 552, pp. 125–143.
- Hannibal, J. Neurotransmitters of the Retino-Hypothalamic Tract. Cell Tissue Res. 2002, 309, 73–88.
- Berson, D.M.; Dunn, F.A.; Takao, M. Phototransduction by Retinal Ganglion Cells That Set the Circadian Clock. Science 2002, 295, 1070–1073.
- Hattar, S.; Liao, H.W.; Takao, M.; Berson, D.M.; Yau, K.W. Melanopsin-Containing Retinal Ganglion Cells: Architecture, Projections, and Intrinsic Photosensitivity. Science 2002, 295, 1065–1070.
- Provencio, I.; Rollag, M.D.; Castrucci, A.M. Photoreceptive Net in the Mammalian Retina. This Mesh of Cells May Explain How Some Blind Mice Can Still Tell Day from Night. Nature 2002, 415, 493.
- Mohawk, J.A.; Green, C.B.; Takahashi, J.S. Central and Peripheral Circadian Clocks in Mammals. Annu. Rev. Neurosci. 2012, 35, 445–462.
- Bunger, M.K.; Wilsbacher, L.D.; Moran, S.M.; Clendenin, C.; Radcliffe, L.A.; Hogenesch, J.B.; Simon, M.C.; Takahashi, J.S.; Bradfield, C.A. Mop3 Is an Essential Component of the Master Circadian Pacemaker in Mammals. Cell 2000, 103, 1009–1017.
- Darlington, T.K.; Wager-Smith, K.; Ceriani, M.F.; Staknis, D.; Gekakis, N.; Steeves, T.D.; Weitz, C.J.; Takahashi, J.S.; Kay, S.A. Closing the Circadian Loop: CLOCK-Induced Transcription of Its Own Inhibitors Per and Tim. Science 1998, 280, 1599–1603.
- Gekakis, N.; Staknis, D.; Nguyen, H.B.; Davis, F.C.; Wilsbacher, L.D.; King, D.P.; Takahashi, J.S.; Weitz, C.J. Role of the CLOCK Protein in the Mammalian Circadian Mechanism. Science 1998, 280, 1564–1569.
- Kume, K.; Zylka, M.J.; Sriram, S.; Shearman, L.P.; Weaver, D.R.; Jin, X.; Maywood, E.S.; Hastings, M.H.; Reppert, S.M. mCRY1 and mCRY2 Are Essential Components of the Negative Limb of the Circadian Clock Feedback Loop. Cell 1999, 98, 193–205.
- Patke, A.; Young, M.W.; Axelrod, S. Molecular Mechanisms and Physiological Importance of Circadian Rhythms. Nat. Rev. Mol. Cell Biol. 2020, 21, 67–84.
- Ye, R.; Selby, C.P.; Chiou, Y.-Y.; Ozkan-Dagliyan, I.; Gaddameedhi, S.; Sancar, A. Dual Modes of CLOCK:BMAL1 Inhibition Mediated by Cryptochrome and Period Proteins in the Mammalian Circadian Clock. Genes Dev. 2014, 28, 1989–1998.
- Ye, R.; Selby, C.P.; Ozturk, N.; Annayev, Y.; Sancar, A. Biochemical Analysis of the Canonical Model for the Mammalian Circadian Clock. J. Biol. Chem. 2011, 286, 25891–25902.
- Cao, X.; Yang, Y.; Selby, C.P.; Liu, Z.; Sancar, A. Molecular Mechanism of the Repressive Phase of the Mammalian Circadian Clock. Proc. Natl. Acad. Sci. USA 2021, 118, e2021174118.
- Jin, J.; Cardozo, T.; Lovering, R.C.; Elledge, S.J.; Pagano, M.; Harper, J.W. Systematic Analysis and Nomenclature of Mammalian F-Box Proteins. Genes Dev. 2004, 18, 2573–2580.
- Eide, E.; Margaret, W.; Heeseog, K.; Peter, W.; William, H.; Fernando, C.; Erica, V.; Andrew, G.; David, V. Control of Mammalian Circadian Rhythm by CKIepsilon-Regulated Proteasome-Mediated PER2 Degradation. Mol. Cell. Biol. 2005, 25, 2795–2807.
- Chen, L.-C.; Hsieh, Y.-L.; Tan, G.Y.T.; Kuo, T.-Y.; Chou, Y.-C.; Hsu, P.-H.; Hwang-Verslues, W.W. Differential Effects of SUMO1 and SUMO2 on Circadian Protein PER2 Stability and Function. Sci. Rep. 2021, 11, 14431.
- Shirogane, T.; Jin, J.; Ang, X.L.; Harper, J.W. SCFβ-TRCP Controls Clock-Dependent Transcription via Casein Kinase 1-Dependent Degradation of the Mammalian Period-1 (Per1) Protein. J. Biol. Chem. 2005, 280, 26863–26872.
- Reischl, S.; Vanselow, K.; Westermark, P.O.; Thierfelder, N.; Maier, B.; Herzel, H.; Kramer, A. β-TrCP1-Mediated Degradation of PERIOD2 Is Essential for Circadian Dynamics. J. Biol. Rhythms 2007, 22, 375–386.
- Ohsaki, K.; Oishi, K.; Kozono, Y.; Nakayama, K.; Nakayama, K.I.; Ishida, N. The Role of β-TrCP1 and β-TrCP2 in Circadian Rhythm Generation by Mediating Degradation of Clock Protein PER2. J. Biochem. 2008, 144, 609–618.
- Masuda, S.; Narasimamurthy, R.; Yoshitane, H.; Kim, J.K.; Fukada, Y.; Virshup, D.M. Mutation of a PER2 Phosphodegron Perturbs the Circadian Phosphoswitch. Proc. Natl. Acad. Sci. USA 2020, 117, 10888–10896.
- D’Alessandro, M.; Beesley, S.; Kim, J.K.; Jones, Z.; Chen, R.; Wi, J.; Kyle, K.; Vera, D.; Pagano, M.; Nowakowski, R.; et al. Stability of Wake-Sleep Cycles Requires Robust Degradation of the PERIOD Protein. Curr. Biol. 2017, 27, 3454–3467.
- Kim, J.; D’Annibale, S.; Magliozzi, R.; Low, T.Y.; Jansen, P.; Shaltiel, I.A.; Mohammed, S.; Heck, A.J.R.; Medema, R.H.; Guardavaccaro, D. USP17- and SCFβTrCP-Regulated Degradation of DEC1 Controls the DNA Damage Response. Mol. Cell. Biol. 2014, 34, 4177–4185.
- Honda, R.; Yasuda, H. Activity of MDM2, a Ubiquitin Ligase, toward P53 or Itself Is Dependent on the RING Finger Domain of the Ligase. Oncogene 2000, 19, 1473–1476.
- Liu, J.; Zou, X.; Gotoh, T.; Brown, A.M.; Jiang, L.; Wisdom, E.L.; Kim, J.K.; Finkielstein, C. V Distinct Control of PERIOD2 Degradation and Circadian Rhythms by the Oncoprotein and Ubiquitin Ligase MDM2. Sci. Signal. 2018, 11, eaau0715.
- Busino, L.; Bassermann, F.; Maiolica, A.; Lee, C.; Nolan, P.M.; Godinho, S.I.H.; Draetta, G.F.; Pagano, M. SCFFbxl3 Controls the Oscillation of the Circadian Clock by Directing the Degradation of Cryptochrome Proteins. Science 2007, 316, 900–904.
- Godinho, S.I.H.; Maywood, E.S.; Shaw, L.; Tucci, V.; Barnard, A.R.; Busino, L.; Pagano, M.; Kendall, R.; Quwailid, M.M.; Romero, M.R.; et al. The After-Hours Mutant Reveals a Role for Fbxl3 in Determining Mammalian Circadian Period. Science 2007, 316, 897–900.
- Siepka, S.M.; Yoo, S.-H.; Park, J.; Song, W.; Kumar, V.; Hu, Y.; Lee, C.; Takahashi, J.S. Circadian Mutant Overtime Reveals F-Box Protein FBXL3 Regulation of Cryptochrome and Period Gene Expression. Cell 2007, 129, 1011–1023.
- Xing, W.; Busino, L.; Hinds, T.R.; Marionni, S.T.; Saifee, N.H.; Bush, M.F.; Pagano, M.; Zheng, N. SCFFBXL3 Ubiquitin Ligase Targets Cryptochromes at Their Cofactor Pocket. Nature 2013, 496, 64–68.
- Yumimoto, K.; Muneoka, T.; Tsuboi, T.; Nakayama, K.I. Substrate Binding Promotes Formation of the Skp1-Cul1-Fbxl3 (SCFFbxl3) Protein Complex. J. Biol. Chem. 2013, 288, 32766–32776.
- Correia, S.; Chan, A.; Vaughan, M.; Zolboot, N.; Perea, V.; Huber, A.-L.; Kriebs, A.; Moresco, J.; Yates, J.; Lamia, K. The Circadian E3 Ligase Complex SCFFBXL3+CRY Targets TLK2. Sci. Rep. 2019, 9, 198.
- Shi, G.; Xing, L.; Liu, Z.; Qu, Z.; Wu, X.; Dong, Z.; Wang, X.; Gao, X.; Huang, M.; Yan, J.; et al. Dual Roles of FBXL3 in the Mammalian Circadian Feedback Loops Are Important for Period Determination and Robustness of the Clock. Proc. Natl. Acad. Sci. USA 2013, 110, 4750–4755.
- Dardente, H.; Mendoza, J.; Fustin, J.-M.; Challet, E.; Hazlerigg, D.G. Implication of the F-Box Protein FBXL21 in Circadian Pacemaker Function in Mammals. PLoS ONE 2008, 3, e3530.
- Yoo, S.-H.; Mohawk, J.A.; Siepka, S.M.; Shan, Y.; Huh, S.K.; Hong, H.-K.; Kornblum, I.; Kumar, V.; Koike, N.; Xu, M.; et al. Competing E3 Ubiquitin Ligases Govern Circadian Periodicity by Degradation of CRY in Nucleus and Cytoplasm. Cell 2013, 152, 1091–1105.
- Hirano, A.; Yumimoto, K.; Tsunematsu, R.; Matsumoto, M.; Oyama, M.; Kozuka-Hata, H.; Nakagawa, T.; Lanjakornsiripan, D.; Nakayama, K.I.; Fukada, Y. FBXL21 Regulates Oscillation of the Circadian Clock through Ubiquitination and Stabilization of Cryptochromes. Cell 2013, 152, 1106–1118.
- Tong, X.; Zhang, D.; Charney, N.; Jin, E.; VanDommelen, K.; Stamper, K.; Gupta, N.; Saldate, J.; Yin, L. DDB1-Mediated CRY1 Degradation Promotes FOXO1-Driven Gluconeogenesis in Liver. Diabetes 2017, 66, 2571–2582.
- Tong, X.; Zhang, D.; Guha, A.; Arthurs, B.; Cazares, V.; Gupta, N.; Yin, L. CUL4-DDB1-CDT2 E3 Ligase Regulates the Molecular Clock Activity by Promoting Ubiquitination-Dependent Degradation of the Mammalian CRY1. PLoS ONE 2015, 10, e0139725.
- Tamayo, A.G.; Duong, H.A.; Robles, M.S.; Mann, M.; Weitz, C.J. Histone Monoubiquitination by Clock–Bmal1 Complex Marks Per1 and Per2 Genes for Circadian Feedback. Nat. Struct. Mol. Biol. 2015, 22, 759–766.
- Okazaki, H.; Matsunaga, N.; Fujioka, T.; Okazaki, F.; Akagawa, Y.; Tsurudome, Y.; Ono, M.; Kuwano, M.; Koyanagi, S.; Ohdo, S. Circadian Regulation of mTOR by the Ubiquitin Pathway in Renal Cell Carcinoma. Cancer Res. 2014, 74, 543–551.
- Mao, J.-H.; Kim, I.-J.; Wu, D.; Climent, J.; Kang, H.C.; DelRosario, R.; Balmain, A. FBXW7 Targets mTOR for Degradation and Cooperates with PTEN in Tumor Suppression. Science 2008, 321, 1499–1502.
- Fang, L.; Yang, Z.; Zhou, J.; Tung, J.-Y.; Hsiao, C.-D.; Wang, L.; Deng, Y.; Wang, P.; Wang, J.; Lee, M.-H. Circadian Clock Gene CRY2 Degradation Is Involved in Chemoresistance of Colorectal Cancer. Mol. Cancer Ther. 2015, 14, 1476–1487.
- Zhao, X.; Hirota, T.; Han, X.; Cho, H.; Chong, L.-W.; Lamia, K.; Liu, S.; Atkins, A.R.; Banayo, E.; Liddle, C.; et al. Circadian Amplitude Regulation via FBXW7-Targeted REV-ERBα Degradation. Cell 2016, 165, 1644–1657.
- Rual, J.-F.; Venkatesan, K.; Hao, T.; Hirozane-Kishikawa, T.; Dricot, A.; Li, N.; Berriz, G.F.; Gibbons, F.D.; Dreze, M.; Ayivi-Guedehoussou, N.; et al. Towards a Proteome-Scale Map of the Human Protein-Protein Interaction Network. Nature 2005, 437, 1173–1178.
- Chen, S.; Yang, J.; Yang, L.; Zhang, Y.; Zhou, L.; Liu, Q.; Duan, C.; Mieres, C.A.; Zhou, G.; Xu, G. Ubiquitin Ligase TRAF2 Attenuates the Transcriptional Activity of the Core Clock Protein BMAL1 and Affects the Maximal Per1 mRNA Level of the Circadian Clock in Cells. FEBS J. 2018, 285, 2987–3001.
- Ullah, K.; Chen, S.; Lu, J.; Wang, X.; Liu, Q.; Zhang, Y.; Long, Y.; Hu, Z.; Xu, G. The E3 Ubiquitin Ligase STUB1 Attenuates Cell Senescence by Promoting the Ubiquitination and Degradation of the Core Circadian Regulator BMAL1. J. Biol. Chem. 2020, 295, 4696–4708.
- Ehlen, J.C.; Jones, K.A.; Pinckney, L.; Gray, C.L.; Burette, S.; Weinberg, R.J.; Evans, J.A.; Brager, A.J.; Zylka, M.J.; Paul, K.N.; et al. Maternal Ube3a Loss Disrupts Sleep Homeostasis But Leaves Circadian Rhythmicity Largely Intact. J. Neurosci. 2015, 35, 13587–13598.
- Kishino, T.; Lalande, M.; Wagstaff, J. UBE3A/E6-AP Mutations Cause Angelman Syndrome. Nat. Genet. 1997, 15, 70–73.
- Jones, K.A.; Han, J.E.; DeBruyne, J.P.; Philpot, B.D. Persistent Neuronal Ube3a Expression in the Suprachiasmatic Nucleus of Angelman Syndrome Model Mice. Sci. Rep. 2016, 6, 28238.
- Shi, S.-Q.; Mahoney, C.E.; Houdek, P.; Zhao, W.; Anderson, M.P.; Zhuo, X.; Beaudet, A.; Sumova, A.; Scammell, T.E.; Johnson, C.H. Circadian Rhythms and Sleep Are Dependent Upon Expression Levels of Key Ubiquitin Ligase Ube3a. Front. Behav. Neurosci. 2022, 16, 837523.
- Thibert, R.L.; Larson, A.M.; Hsieh, D.T.; Raby, A.R.; Thiele, E.A. Neurologic Manifestations of Angelman Syndrome. Pediatr. Neurol. 2013, 48, 271–279.
- Colas, D.; Wagstaff, J.; Fort, P.; Salvert, D.; Sarda, N. Sleep Disturbances in Ube3a Maternal-Deficient Mice Modeling Angelman Syndrome. Neurobiol. Dis. 2005, 20, 471–478.
- Shi, S.; Bichell, T.J.; Ihrie, R.A.; Johnson, C.H. Ube3a Imprinting Impairs Circadian Robustness in Angelman Syndrome Models. Curr. Biol. 2015, 25, 537–545.
- Gossan, N.C.; Zhang, F.; Guo, B.; Jin, D.; Yoshitane, H.; Yao, A.; Glossop, N.; Zhang, Y.Q.; Fukada, Y.; Meng, Q.-J. The E3 Ubiquitin Ligase UBE3A Is an Integral Component of the Molecular Circadian Clock through Regulating the BMAL1 Transcription Factor. Nucleic Acids Res. 2014, 42, 5765–5775.
- Ullah, K.; Zubia, E.; Narayan, M.; Yang, J.; Xu, G. Diverse Roles of the E2/E3 Hybrid Enzyme UBE2O in the Regulation of Protein Ubiquitination, Cellular Functions, and Disease Onset. FEBS J. 2019, 286, 2018–2034.
- Chen, S.; Yang, J.; Zhang, Y.; Duan, C.; Liu, Q.; Huang, Z.; Xu, Y.; Zhou, L.; Xu, G. Ubiquitin-Conjugating Enzyme UBE2O Regulates Cellular Clock Function by Promoting the Degradation of the Transcription Factor BMAL1. J. Biol. Chem. 2018, 293, 11296–11309.
- DeBruyne, J.P.; Baggs, J.E.; Sato, T.K.; Hogenesch, J.B. Ubiquitin Ligase Siah2 Regulates RevErbα Degradation and the Mammalian Circadian Clock. Proc. Natl. Acad. Sci. USA 2015, 112, 12420–12425.
- Mekbib, T.; Suen, T.-C.; Rollins-Hairston, A.; Smith, K.; Armstrong, A.; Gray, C.; Owino, S.; Baba, K.; Baggs, J.E.; Ehlen, J.C.; et al. The Ubiquitin Ligase SIAH2 Is a Female-Specific Regulator of Circadian Rhythms and Metabolism. PLoS Genet. 2022, 18, e1010305.
- Chen, D.; Brooks, C.L.; Gu, W. ARF-BP1 as a Potential Therapeutic Target. Br. J. Cancer 2006, 94, 1555–1558.
- Han, S.; Witt, R.M.; Santos, T.M.; Polizzano, C.; Sabatini, B.L.; Ramesh, V. Pam (Protein Associated with Myc) Functions as an E3 Ubiquitin Ligase and Regulates TSC/mTOR Signaling. Cell. Signal. 2008, 20, 1084–1091.
- Yin, L.; Joshi, S.; Wu, N.; Tong, X.; Lazar, M.A. E3 Ligases Arf-Bp1 and Pam Mediate Lithium-Stimulated Degradation of the Circadian Heme Receptor Rev-Erbα. Proc. Natl. Acad. Sci. USA 2010, 107, 11614–11619.
