MicroRNAs (miRNAs) are small noncoding RNAs that play an important role in regulating gene expression at a posttranscriptional level. As one of the first discovered oncogenic miRNAs, microRNA-21 (miR-21) has been highlighted for its critical role in cancers, such as glioblastoma, pancreatic adenocarcinoma, non-small cell lung cancer, and many others. MiR-21 targets many vital components in a wide range of cancers and acts on various cellular processes ranging from cancer stemness to cell death. Expression of miR-21 is elevated within cancer tissues and circulating miR-21 is readily detectable in biofluids, making it valuable as a cancer biomarker with significant potential for use in diagnosis and prognosis.
- noncoding RNA
- microRNA-21
- cancer
- biomarker
- RNA therapeutics
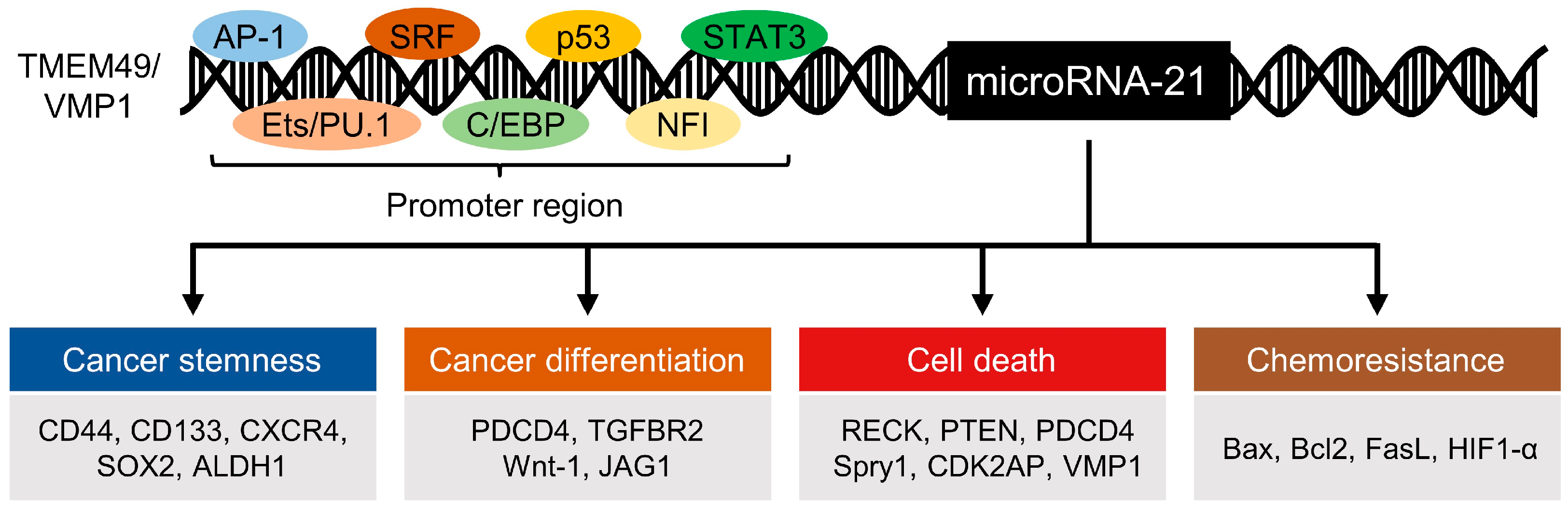
1. MicroRNA-21 and Its Role in Cancer
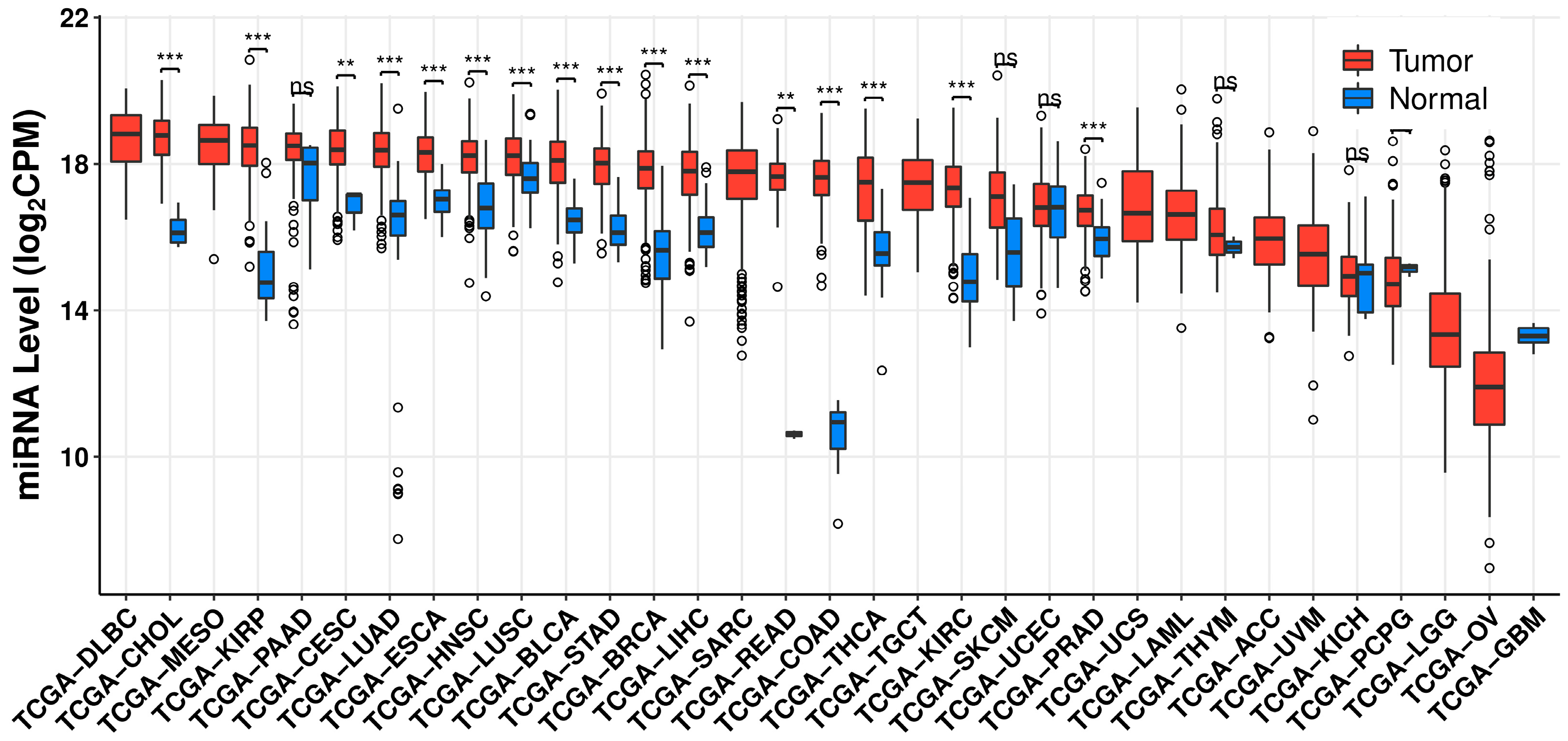
2. MicroRNA-21 as a Biomarker in Cancer
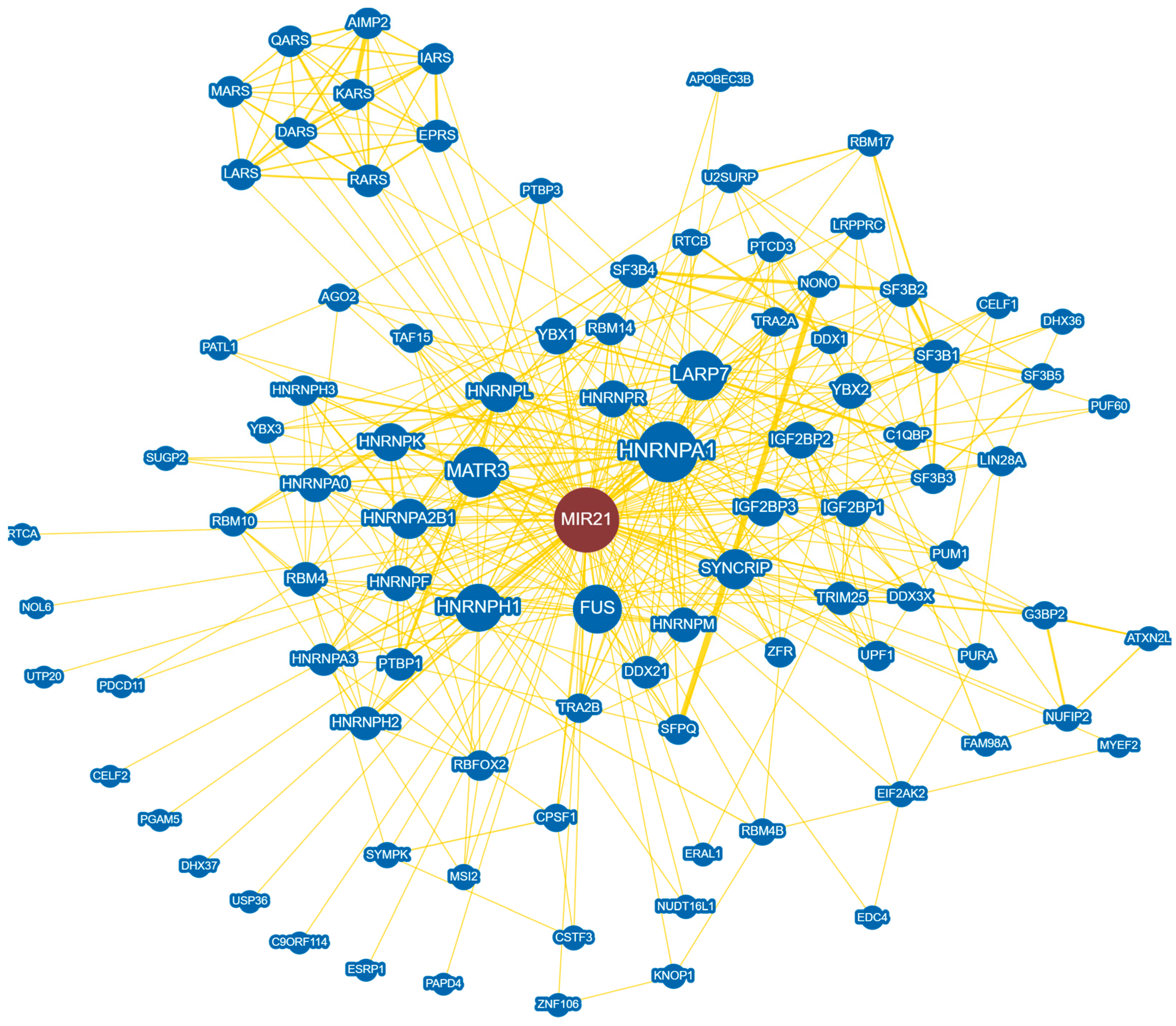
3. MicroRNA-21-Targeted RNA Therapeutics
| Type | Target Cancers | Cell Lines & Experimental Models |
Phenotypes | References |
|---|---|---|---|---|
| LNA | Colorectal adenocarcinoma |
LS174T | Proliferation inhibited; apoptosis enhanced |
[46] |
| Melanoma | B16F10; C57BL/6 mice |
Tumor growth and volume inhibited; apoptosis enhanced |
[47] | |
| Glioblastoma | U87MG; Orthotopic xenograft in athymic nude mice |
Tumor growth inhibited; apoptosis enhanced |
[48][49] | |
| Non-small cell lung cancer | A549; Female nude mice |
Drug sensitization; tumor growth inhibited; apoptosis enhanced |
[50] | |
| Breast cancer | MCF-7 | Proliferation inhibited | [23] | |
| ASO | Laryngeal squamous cell carcinoma |
Hep-2; BALB/c nude mice |
Tumor growth and proliferation inhibited; invasiveness decreased; cell cycle arrest; apoptosis enhanced |
[51] |
| Non-small cell lung cancer | PC9; Female BALB/c nude mice |
Proliferation inhibited; apoptosis enhanced; tumor growth inhibited |
[52] | |
| Glioblastoma multiforme |
LN229; U251MG; U373MG; T98G |
Drug sensitization; cell viability decreased |
[53][54] | |
| Hepatocellular carcinoma |
Huh7; HepG2 | Migration and invasiveness decreased |
[55] | |
| Breast phyllode tumor |
Patient-derived breast stromal cells; Female nude mice |
Proliferation inhibited and invasion decreased |
[56] | |
| CircRNA | Gastric carcinoma | NCI-N87; AGS; MKN28 | Proliferation inhibited | [57] |
| Lung cancer | L132, A549; LL2; 3D multicellular spheroids |
Proliferation inhibited; migration decreased; apoptosis enhanced |
[58] |
This entry is adapted from the peer-reviewed paper 10.3390/cells11182791
References
- Fujita, S.; Ito, T.; Mizutani, T.; Minoguchi, S.; Yamamichi, N.; Sakurai, K.; Iba, H. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J. Mol. Biol. 2008, 378, 492–504.
- Feng, Y.H.; Tsao, C.J. Emerging role of microRNA-21 in cancer. Biomed. Rep. 2016, 5, 395–402.
- Bailey, M.H.; Tokheim, C.; Porta-Pardo, E.; Sengupta, S.; Bertrand, D.; Weerasinghe, A.; Colaprico, A.; Wendl, M.C.; Kim, J.; Reardon, B.; et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 2018, 173, 371–385.
- Zhou, Y.; Guo, D.; Zhang, Y. Association of MicroRNA-21 with p53 at Mutant Sites R175H and R248Q, Clinicopathological Features, and Prognosis of NSCLC. Mol. Ther. Oncolytics 2020, 19, 208–217.
- Bornachea, O.; Santos, M.; Martínez-Cruz, A.B.; García-Escudero, R.; Dueñas, M.; Costa, C.; Segrelles, C.; Lorz, C.; Buitrago, A.; Saiz-Ladera, C.; et al. EMT and induction of miR-21 mediate metastasis development in Trp53-deficient tumours. Sci. Rep. 2012, 2, 434.
- Chu, N.J.; Anders, R.A.; Fertig, E.J.; Cao, M.; Hopkins, A.C.; Keenan, B.P. Inhibition of miR-21 Regulates Mutant KRAS Effector Pathways and Intercepts Pancreatic Ductal Adenocarcinoma Development. Cancer Prev. Res. 2020, 13, 569–582.
- Hatley, M.E.; Patrick, D.M.; Garcia, M.R.; Richardson, J.A.; Bassel-Duby, R.; van Rooij, E.; Olson, E.N. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell 2010, 18, 282–293.
- Seike, M.; Goto, A.; Okano, T.; Bowman, E.D.; Schetter, A.J.; Horikawa, I.; Mathe, E.A.; Jen, J.; Yang, P.; Sugimura, H.; et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc. Natl. Acad. Sci. USA 2009, 106, 12085–12090.
- Shen, J.; Xia, W.; Khotskaya, Y.B.; Huo, L.; Nakanishi, K.; Lim, S.O.; Du, Y.; Wang, Y.; Chang, W.C.; Chen, C.H.; et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature 2013, 497, 383–387.
- Bloomston, M.; Frankel, W.L.; Petrocca, F.; Volinia, S.; Alder, H.; Hagan, J.P.; Liu, C.G.; Bhatt, D.; Taccioli, C.; Croce, C.M. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA 2007, 297, 1901–1908.
- Iorio, M.V.; Ferracin, M.; Liu, C.G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005, 65, 7065–7070.
- Schetter, A.J.; Leung, S.Y.; Sohn, J.J.; Zanetti, K.A.; Bowman, E.D.; Yanaihara, N.; Yuen, S.T.; Chan, T.L.; Kwong, D.L.; Au, G.K.; et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 2008, 299, 425–436.
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261.
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129, 1401–1414.
- Ciafrè, S.A.; Galardi, S.; Mangiola, A.; Ferracin, M.; Liu, C.G.; Sabatino, G.; Negrini, M.; Maira, G.; Croce, C.M.; Farace, M.G. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 2005, 334, 1351–1358.
- Li, R.; Qu, H.; Wang, S. CancerMIRNome: An interactive analysis and visualization database for miRNome profiles of human cancer. Nucleic Acids Res. 2022, 50, D1139–D1146.
- Ren, Y.; Zhou, X.; Mei, M.; Yuan, X.B.; Han, L.; Wang, G.X.; Jia, Z.F.; Xu, P.; Pu, P.Y.; Kang, C.S. MicroRNA-21 inhibitor sensitizes human glioblastoma cells U251 (PTEN-mutant) and LN229 (PTEN-wild type) to taxol. BMC Cancer 2010, 10, 27.
- Han, L.; Yue, X.; Zhou, X.; Lan, F.M.; You, G.; Zhang, W.; Zhang, K.L.; Zhang, C.Z.; Cheng, J.Q.; Yu, S.Z.; et al. MicroRNA-21 expression is regulated by β-catenin/STAT3 pathway and promotes glioma cell invasion by direct targeting RECK. CNS Neurosci. Ther. 2012, 18, 573–583.
- Zhang, K.L.; Han, L.; Chen, L.Y.; Shi, Z.D.; Yang, M.; Ren, Y.; Chen, L.C.; Zhang, J.X.; Pu, P.Y.; Kang, C.S. Blockage of a miR-21/EGFR regulatory feedback loop augments anti-EGFR therapy in glioblastomas. Cancer Lett. 2014, 342, 139–149.
- Kwak, H.J.; Kim, Y.J.; Chun, K.R.; Woo, Y.M.; Park, S.J.; Jeong, J.A.; Jo, S.H.; Kim, T.H.; Min, H.S.; Chae, J.S.; et al. Downregulation of Spry2 by miR-21 triggers malignancy in human gliomas. Oncogene 2011, 30, 2433–2442.
- Zhao, Q.; Chen, S.; Zhu, Z.; Yu, L.; Ren, Y.; Jiang, M.; Weng, J. miR-21 promotes EGF-induced pancreatic cancer cell proliferation by targeting Spry2. Cell Death Dis. 2018, 9, 1157.
- Zhang, J.G.; Wang, J.J.; Zhao, F.; Liu, Q.; Jiang, K.; Yang, G.H. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC). Clin. Chim. Acta 2010, 411, 846–852.
- Frankel, L.B.; Christoffersen, N.R.; Jacobsen, A.; Lindow, M.; Krogh, A.; Lund, A.H. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J. Biol. Chem. 2008, 283, 1026–1033.
- Zhu, S.; Wu, H.; Wu, F.; Nie, D.; Sheng, S.; Mo, Y.Y. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008, 18, 350–359.
- Medina, P.P.; Nolde, M.; Slack, F.J. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 2010, 467, 86–90.
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297.
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011, 8, 467–477.
- Creemers, E.E.; Tijsen, A.J.; Pinto, Y.M. Circulating microRNAs: Novel biomarkers and extracellular communicators in cardiovascular disease? Circ. Res. 2012, 110, 483–495.
- Tomimaru, Y.; Eguchi, H.; Nagano, H.; Wada, H.; Kobayashi, S.; Marubashi, S.; Tanemura, M.; Tomokuni, A.; Takemasa, I.; Umeshita, K.; et al. Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. J. Hepatol. 2012, 56, 167–175.
- Sierzega, M.; Kaczor, M.; Kolodziejczyk, P.; Kulig, J.; Sanak, M.; Richter, P. Evaluation of serum microRNA biomarkers for gastric cancer based on blood and tissue pools profiling: The importance of miR-21 and miR-331. Br. J. Cancer 2017, 117, 266–273.
- Guo, X.; Lv, X.; Lv, X.; Ma, Y.; Chen, L.; Chen, Y. Circulating miR-21 serves as a serum biomarker for hepatocellular carcinoma and correlated with distant metastasis. Oncotarget 2017, 8, 44050–44058.
- Qu, K.Z.; Zhang, K.; Li, H.; Afdhal, N.H.; Albitar, M. Circulating microRNAs as biomarkers for hepatocellular carcinoma. J. Clin. Gastroenterol. 2011, 45, 355–360.
- Lee, K.-Y.; Seo, Y.; Im, J.H.; Rhim, J.; Baek, W.; Kim, S.; Kwon, J.-W.; Yoo, B.C.; Shin, S.H.; Yoo, H.; et al. Molecular Signature of Extracellular Vesicular Small Non-Coding RNAs Derived from Cerebrospinal Fluid of Leptomeningeal Metastasis Patients: Functional Implication of miR-21 and Other Small RNAs in Cancer Malignancy. Cancers 2021, 13, 209.
- Im, J.H.; Yoo, B.C.; Lee, J.H.; Lee, K.-Y.; Kim, K.-H.; Kim, J.H.; Park, H.J.; Park, M.; Lee, S.H.; Kwon, J.-W.; et al. Experimental Assessment of Leptomeningeal Metastasis Diagnosis in Medulloblastoma Using Cerebrospinal Fluid Metabolomic Profiles. Metabolites 2021, 11, 851.
- Toiyama, Y.; Takahashi, M.; Hur, K.; Nagasaka, T.; Tanaka, K.; Inoue, Y.; Kusunoki, M.; Boland, C.R.; Goel, A. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J. Natl. Cancer Inst. 2013, 105, 849–859.
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21.
- Lee, K.Y.; Im, J.H.; Lin, W.; Gwak, H.S. Nanoparticles in 472 Human Cerebrospinal Fluid: Changes in Extracellular Vesicle Concentration and miR-21 Expression as a Biomarker for Leptomeningeal Metastasis. Cancers 2020, 12, 2745.
- Im, J.H.; Kim, T.H.; Lee, K.-Y.; Gwak, H.-S.; Lin, W.; Park, J.B.; Kim, J.H.; Yoo, B.C.; Park, S.-M.; Kwon, J.-W.; et al. Exploratory Profiling of Extracellular MicroRNAs in Cerebrospinal Fluid Comparing Leptomeningeal Metastasis with Other Central Nervous System Tumor Statuses. J. Clin. Med. 2021, 10, 4860.
- Im, J.H.; Yoo, B.C.; Lee, J.H.; Kim, K.H.; Kim, T.H.; Lee, K.Y.; Kim, J.H.; Park, J.B.; Kwon, J.W.; Shin, S.H.; et al. Comparative cerebrospinal fluid metabolites profiling in glioma patients to predict malignant transformation and leptomeningeal metastasis with a potential for preventive personalized medicine. EPMA J. 2020, 11, 469–484.
- Li, J.R.; Tong, C.Y.; Sung, T.J.; Kang, T.Y.; Zhou, X.J.; Liu, C.C. CMEP: A database for circulating microRNA expression profiling. Bioinformatics 2019, 35, 3127–3132.
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.J.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021, 30, 187–200.
- Stark, C.; Breitkreutz, B.J.; Reguly, T.; Boucher, L.; Breitkreutz, A.; Tyers, M. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 2006, 34, D535–D539.
- López-Urrutia, E.; Bustamante Montes, L.P.; Ladrón de Guevara Cervantes, D.; Pérez-Plasencia, C.; Campos-Parra, A.D. Crosstalk Between Long Non-coding RNAs, Micro-RNAs and mRNAs: Deciphering Molecular Mechanisms of Master Regulators in Cancer. Front. Oncol. 2019, 9, 669.
- Jeggari, A.; Marks, D.S.; Larsson, E. miRcode: A map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics 2012, 28, 2062–2063.
- Seo, Y.; Kim, S.S.; Kim, N.; Cho, S. Development of a miRNA-controlled dual-sensing system and its application for targeting miR-21 signaling in tumorigenesis. Exp. Mol. Med. 2020, 52, 1989–2004.
- Nedaeinia, R.; Sharifi, M. Locked nucleic acid anti-miR-21 inhibits cell growth and invasive behaviors of a colorectal adenocarcinoma cell line: LNA-anti-miR as a novel approach. Cancer Gene Ther. 2016, 23, 246–253.
- Javanmard, S.H.; Vaseghi, G.; Ghasemi, A.; Rafiee, L.; Ferns, G.A.; Esfahani, H.N.; Nedaeinia, R. Therapeutic inhibition of microRNA-21 (miR-21) using locked-nucleic acid (LNA)-anti-miR and its effects on the biological behaviors of melanoma cancer cells in preclinical studies. Cancer Cell Int. 2020, 20, 384.
- Lee, T.J.; Yoo, J.Y.; Shu, D.; Li, H.; Zhang, J.; Yu, J.G.; Jaime-Ramirez, A.C.; Acunzo, M.; Romano, G.; Cui, R.; et al. RNA Nanoparticle-Based Targeted Therapy for Glioblastoma through Inhibition of Oncogenic miR-21. Mol. Ther. 2017, 25, 1544–1555.
- Griveau, A.; Bejaud, J.; Anthiya, S.; Avril, S.; Autret, D.; Garcion, E. Silencing of miR-21 by locked nucleic acid-lipid nanocapsule complexes sensitize human glioblastoma cells to radiation-induced cell death. Int. J. Pharm. 2013, 454, 765–774.
- Xu, L.; Huang, Y.; Chen, D.; He, J.; Zhu, W.; Zhang, Y.; Liu, X. Downregulation of miR-21 increases cisplatin sensitivity of non-small-cell lung cancer. Cancer Genet. 2014, 207, 214–220.
- Ren, J.; Zhu, D.; Liu, M.; Sun, Y.; Tian, L. Downregulation of miR-21 modulates Ras expression to promote apoptosis and suppress invasion of Laryngeal squamous cell carcinoma. Eur. J. Cancer 2010, 46, 3409–3416.
- Li, B.; Ren, S.; Li, X.; Wang, Y.; Garfield, D.; Zhou, S.; Chen, X.; Su, C.; Chen, M.; Kuang, P.; et al. MiR-21 overexpression is associated with acquired resistance of EGFR-TKI in non-small cell lung cancer. Lung Cancer 2014, 83, 146–153.
- Li, Y.; Li, W.; Yang, Y.; Lu, Y.; He, C.; Hu, G.; Liu, H.; Chen, J.; He, J.; Yu, H. MicroRNA-21 targets LRRFIP1 and contributes to VM-26 resistance in glioblastoma multiforme. Brain Res. 2009, 1286, 13–18.
- Giunti, L.; da Ros, M.; Vinci, S.; Gelmini, S.; Iorio, A.L.; Buccoliero, A.M.; Cardellicchio, S.; Castiglione, F.; Genitori, L.; de Martino, M.; et al. Anti-miR21 oligonucleotide enhances chemosensitivity of T98G cell line to doxorubicin by inducing apoptosis. Am. J. Cancer Res. 2015, 5, 231–242.
- Connolly, E.C.; Van Doorslaer, K.; Rogler, L.E.; Rogler, C.E. Overexpression of miR-21 promotes an in vitro metastatic phenotype by targeting the tumor suppressor RHOB. Mol. Cancer Res. 2010, 8, 691–700.
- Gong, C.; Nie, Y.; Qu, S.; Liao, J.Y.; Cui, X.; Yao, H.; Zeng, Y.; Su, F.; Song, E.; Liu, Q. miR-21 induces myofibroblast differentiation and promotes the malignant progression of breast phyllodes tumors. Cancer Res. 2014, 74, 4341–4352.
- Liu, X.; Abraham, J.M.; Cheng, Y.; Wang, Z.; Wang, Z.; Zhang, G.; Ashktorab, H.; Smoot, D.T.; Cole, R.N.; Boronina, T.N.; et al. Synthetic Circular RNA Functions as a miR-21 Sponge to Suppress Gastric Carcinoma Cell Proliferation. Mol. Ther. Nucleic Acids 2018, 13, 312–321.
- Rama, A.R.; Quiñonero, F. Synthetic Circular miR-21 Sponge as Tool for Lung Cancer Treatment. Int. J. Mol. Sci. 2022, 23, 2963.
- Davis, S.; Lollo, B.; Freier, S.; Esau, C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006, 34, 2294–2304.
- Burris, H.A., 3rd; Moore, M.J.; Andersen, J.; Green, M.R.; Rothenberg, M.L.; Modiano, M.R.; Cripps, M.C.; Portenoy, R.K.; Storniolo, A.M.; Tarassoff, P.; et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: A randomized trial. J. Clin. Oncol. 1997, 15, 2403–2413.
- Li, Y.; Chen, Y.; Li, J.; Zhang, Z.; Huang, C.; Lian, G.; Yang, K.; Chen, S.; Lin, Y.; Wang, L.; et al. Co-delivery of microRNA-21 antisense oligonucleotides and gemcitabine using nanomedicine for pancreatic cancer therapy. Cancer Sci. 2017, 108, 1493–1503.
- Si, M.L.; Zhu, S.; Wu, H.; Lu, Z.; Wu, F.; Mo, Y.Y. miR-21-mediated tumor growth. Oncogene 2007, 26, 2799–2803.
- Leone, E.; Morelli, E.; Di Martino, M.T.; Amodio, N.; Foresta, U.; Gullà, A.; Rossi, M.; Neri, A.; Giordano, A.; Munshi, N.C.; et al. Targeting miR-21 inhibits in vitro and in vivo multiple myeloma cell growth. Clin. Cancer Res. 2013, 19, 2096–2106.
- Wagenaar, T.R.; Tolstykh, T.; Shi, C.; Jiang, L.; Zhang, J.; Li, Z.; Yu, Q.; Qu, H.; Sun, F.; Cao, H.; et al. Identification of the endosomal sorting complex required for transport-I (ESCRT-I) as an important modulator of anti-miR uptake by cancer cells. Nucleic Acids Res. 2015, 43, 1204–1215.
- Zhou, X.; Zhang, J.; Jia, Q.; Ren, Y.; Wang, Y.; Shi, L.; Liu, N.; Wang, G.; Pu, P.; You, Y.; et al. Reduction of miR-21 induces glioma cell apoptosis via activating caspase 9 and 3. Oncol. Rep. 2010, 24, 195–201.
- Koshkin, A.A.; Singh, S.K.; Nielsen, P.; Rajwanshi, V.K.; Kumar, R.; Meldgaard, M.; Olsen, C.E.; Wengel, J. LNA (Locked Nucleic Acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron 1998, 54, 3607–3630.
- Lima, J.F.; Cerqueira, L. Anti-miRNA oligonucleotides: A comprehensive guide for design. RNA Biol. 2018, 15, 338–352.
- Obad, S.; dos Santos, C.O.; Petri, A.; Heidenblad, M.; Broom, O.; Ruse, C.; Fu, C.; Lindow, M.; Stenvang, J.; Straarup, E.M.; et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat. Genet. 2011, 43, 371–378.
- Du, W.W.; Zhang, C.; Yang, W.; Yong, T.; Awan, F.M.; Yang, B.B. Identifying and Characterizing circRNA-Protein Interaction. Theranostics 2017, 7, 4183–4191.
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264.
- Zhong, Y.; Du, Y.; Yang, X.; Mo, Y.; Fan, C.; Xiong, F.; Ren, D.; Ye, X.; Li, C.; Wang, Y.; et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol. Cancer 2018, 17, 79.
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-coding RNA networks in cancer. Nat. Rev. Cancer 2018, 18, 5–18.
- Ebert, M.S.; Neilson, J.R.; Sharp, P.A. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 2007, 4, 721–726.
- Jung, J.; Yeom, C.; Choi, Y.S.; Kim, S.; Lee, E.; Park, M.J.; Kang, S.W.; Kim, S.B.; Chang, S. Simultaneous inhibition of multiple oncogenic miRNAs by a multi-potent microRNA sponge. Oncotarget 2015, 6, 20370–20387.
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388.
- Zhang, C.C.; Li, Y.; Feng, X.Z.; Li, D.B. Circular RNA circ_0001287 inhibits the proliferation, metastasis, and radiosensitivity of non-small cell lung cancer cells by sponging microRNA miR-21 and up-regulating phosphatase and tensin homolog expression. Bioengineered 2021, 12, 414–425.
- Müller, S.; Wedler, A.; Breuer, J.; Glaß, M.; Bley, N.; Lederer, M.; Haase, J.; Misiak, C.; Fuchs, T.; Ottmann, A.; et al. Synthetic circular miR-21 RNA decoys enhance tumor suppressor expression and impair tumor growth in mice. NAR Cancer 2020, 2, zcaa014.
- Wang, X.; Yang, T.; Yu, Z.; Liu, T.; Jin, R.; Weng, L.; Bai, Y.; Gooding, J.J.; Zhang, Y.; Chen, X. Intelligent Gold Nanoparticles with Oncogenic MicroRNA-Dependent Activities to Manipulate Tumorigenic Environments for Synergistic Tumor Therapy. Adv. Mater. 2022, 34, e2110219.
