Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Los coronavirus son una gran familia de patógenos bien establecidos de varios huéspedes, incluidos animales domésticos, animales salvajes y humanos.
- lateral flow immunoassay
- biosensors
- COVID-19
1. Descripción general del SARS-CoV-2
Los coronavirus son una gran familia de patógenos bien establecidos de varios huéspedes, incluidos animales domésticos, animales salvajes y humanos [ 1 ] . Los virus que causaron brotes anteriores en humanos, causando enfermedades respiratorias graves, lesiones pulmonares y la muerte, son el SARS-CoV (coronavirus del síndrome respiratorio agudo severo) en 2003 y el MERS-CoV (coronavirus del síndrome respiratorio de Oriente Medio) en 2012 [ 2 ] . Un análisis genómico reciente con varias herramientas bioinformáticas demostró que el SARS-CoV-2 tiene un genoma muy similar al del coronavirus Bat y el dominio de unión al receptor (RBD) de la glicoproteína espiga como el coronavirus del pangolín malayo [ 3 ]. Esta evidencia indica que el murciélago de herradura es el reservorio natural, y la evidencia principal sugiere que el pangolín malayo es un huésped intermediario [ 3 ] .
El SARS-CoV-2 es un virus envuelto con un ARN monocatenario de sentido positivo. El tamaño del genoma de este patógeno varía de 29,8 kb a 29,9 kb [ 4 ] . El virus codifica al menos 29 proteínas. Las proteínas estructurales son proteínas de espiga (S), membrana (M), envoltura (E) y nucleocápside (NP) [ 5 ] . Las proteínas no estructurales (nsps) tienen funciones necesarias para la replicación y transcripción en el ciclo de vida del virus [ 6 ] . El tamaño de las partículas virales oscila entre 80 y 120 nm [ 7 ] .
El mecanismo de infección viral en humanos es a través de gotitas y aerosoles, que pueden viajar por el aire [ 8 ] . La infección ocurre en células que expresan ACE2 (enzima convertidora de angiotensina 2) y TMPRSS2 (proteasa de serina transmembrana 2) [ 9 ] . La proteína S del coronavirus se une a ACE2, el principal receptor del SARS-CoV-2 que media la entrada del virus en las células, y TMPRSS2 escinde la proteína S (en las subunidades S1 y S2) del SARS-CoV-2, lo que facilita la fusión del SARS- CoV-2 y membrana celular [ 9 ] [ 10 ] [ 11 ]. Además, se ha demostrado que las cisteína proteasas catepsina B y catepsina L endosomal también pueden contribuir a este proceso [ 10 ] [ 12 ] [ 13 ] . En el tracto respiratorio, ACE2 y TMPRSS2 se expresan en las células secretoras y ciliadas de la nariz, las células secretoras y ciliadas de las vías respiratorias conductoras, en las células alveolares de tipo II en los pulmones y en la conjuntiva corneal del ojo [ 14 ] [ 15 ] [ 16 ] [ 17 ] .
The etiological virus of the pandemic has continuously evolved, with many variants emerging worldwide. Variants are categorized as the variant of interest, variant of concern, and variant under monitoring [18]. There are five SARS-CoV-2 lineages designated as the variant of concern alpha, beta, gamma, delta, and omicron variants [19]. These variants increase transmissibility compared to the original virus and potentially increase disease severity [20].
2. Immune Response against SARS-CoV-2 in Brief
The SARS-CoV-2 infection involves diverse stages in the individual: start of infection, disease development, recovery, or systemic compromise. Each infection stage triggers and modulates innate and adaptative immune system mechanisms. Although SARS-CoV-2 is a virus that humanity is learning about, the immune response is equipped with mechanisms capable of dealing with this new threat. In the initial phase of SARS-CoV-2 infection, the individual presents a presymptomatic phase lasting up to 5 days, in which a high viral load is present [21]. In these early days of infection, antibodies may not have been produced. Therefore, innate immunity is the first activated. The innate immune response comprises soluble and cellular components that respond nonspecifically against the virus. The cellular compounds include dendritic cells (DC), monocytes, macrophages, neutrophils, natural killer (NK) cells, and other innate lymphoid cells (ILCs) [22]. Whereas soluble components include complement systems, soluble proteins, interferons, chemokines, and naturally occurring antibodies [23]. Immune response cells recognize pathogen-associated molecular patterns (PAMPs) of SARS-CoV-2 through pattern recognition receptors (PRRs) such as Toll-like receptors (TLR), RIG-I-like receptors (RLR), and melanoma differentiation-associated protein 5 (MDA5). The viral sensing triggers the activation of signaling pathways which induce the production of immune mediators to generate an antiviral state mainly mediated by type I (IFN-α/β) and type III (IFN-λ) interferons (IFNs) [24]. Reports have described that robust IFNs production during the early stage of infection is required to have a protective innate immune response against the virus [25]. On the contrary, an inadequate and slow response to type I and type III IFNs due to virus evasion mechanisms, host comorbidities, or genetic defects cause an exacerbated immune response. This inadequate response induces elevated levels of chemokines (CCL2, CCL8, CXCL2, CXCL8, CXCL9, and CXCL16), high expression of proinflammatory cytokines such as IL-6, IL-10, IL-1, and TNF, in addition to activation, and recruitment of immune cells [26][27]. The called “cytokine storm” leads to unbalanced levels of proinflammatory and antiviral mediators that remain the leading cause of ARDS and multi-organ failure [25][26][28].
On the other hand, the adaptive immune response is orchestrated by CD8+ T lymphocytes, TCD4+, and B lymphocytes, responsible for immunological memory. In response to SARS-CoV-2 infection, it has been shown that non-severe patients or patients with mild symptoms have a low viral load and may not have produced antibodies [29][30]. In contrast, antibodies have been detected by immunoassay tests and biosensors in patients with severe symptoms or cases [29][31]. Patients with a high viral load activate the humoral immune response in the first two weeks of infection [32]. The first seroconversion of antibodies is against protein N, followed by protein S of SARS-CoV-2 in patients with disease symptoms [33]. Immunoglobulins IgA and IgM begin to be detected within the first ten days of infection; however, both antibodies can cross-react with protein N, which is highly conserved among coronaviruses [34][35]. Moreover, high levels of IgG1 and IgG3 are expressed ten to fourteen days after infection in patients with disease symptoms [36][37]. Older adults and seriously ill individuals reach high specificity antibodies concentrations against SARS-CoV-2 S protein.
Due to the urgency of reducing thousands of people’s cases and deaths, scientists have developed several vaccines against COVID-19. Efforts are being made to apply vaccines with emergency use authorization to the world population. Vaccination elicits immune responses capable of potently neutralizing SARS-CoV-2. However, the available data show that most approved COVID-19 vaccines protect against severe disease but do not prevent the clinical manifestation of COVID-19 [38]. Instead, it has been demonstrated that new variants with mutations in the spike, the main target of neutralizing antibodies, can escape the neutralization of humoral immunity [39][40].
3. SARS-CoV-2 Detection
Molecular tests or biosensors are the tools for detecting SARS-CoV-2 nucleic acids/ antigens/antibodies against the virus (Figure 1). In the early part of the illness, viral particles and their subunits can be detected; beyond the first two weeks of illness onset, antibodies against the virus could be detected [41]. The SARS-CoV-2 infection stage is highly correlated to the diagnostic technique recommended for the pandemic. Early diagnosis of the disease and isolation of infected people is key to controlling the transmission of SARS-CoV-2 [42][43]. In the initial phase of SARS-CoV-2 infection, the individual presents a presymptomatic phase lasting up to 5 days, in which a high viral load is present [21]. During these early days of infection, antibodies may not be detected. Therefore, since the pandemic began, the diagnostic method has been based on detecting viral genes using the molecular PCR technique, the gold standard worldwide [44][45][46]. The pandemic has exceeded the ability to identify the virus in laboratories using molecular techniques; this has motivated the development of new technologies for the rapid detection of SARS-CoV-2 that are easy to perform compared to molecular tests in clinical laboratories. LFIA has been the unique device approved and available to use in mass worldwide. Biosensors with transducers are developing in SARS-CoV-2 diagnostic. However, most nanomaterials used in these biosensors present interferences with contaminants in human samples compared to performance under experimental conditions. It is important to emphasize that LFIAs have the unique properties of availability, accessibility, economy, and POC (including home use), these characteristics that are not shared by all biosensors with a transducer. In addition, biosensors with transducers require exclusive handling in laboratories certified under the Clinical Laboratory Improvement Amendments of 1998 [47][48]. The FDA have to date approved only one piezoelectric biosensor [47] (Figure 1).
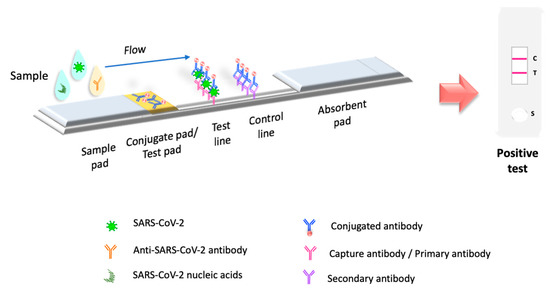
Figure 1. Principio de la prueba LFIA. La prueba LFIA detecta la molécula objetivo en una membrana absorbente con anticuerpos alineados para formar las líneas de prueba y control. La muestra se coloca en una almohadilla de muestra, luego migra a la almohadilla de conjugado, que contiene el conjugado inmovilizado, generalmente hecho de nanopartículas (oro coloidal, látex coloreado o fluorescente, celulosa coloreada) conjugadas con anticuerpos o antígenos. La muestra interactúa con el conjugado y ambos migran a la siguiente sección de la tira, donde se inmovilizan los componentes biológicos del ensayo (proteínas/anticuerpos/antígenos). En esta sección, se capturan el analito y el conjugado. El exceso de reactivo pasa a través de las líneas de captura y se acumula en la almohadilla absorbente. Los resultados se interpretan en la membrana de nitrocelulosa como la presencia o ausencia de las líneas de prueba y control.
This entry is adapted from the peer-reviewed paper 10.3390/bios12090728
References
- Hasöksüz, M.; Kiliç, S.; Saraç, F. Coronaviruses and SARS-COV-2. Turk. J. Med. Sci. 2020, 50, 549–556.
- Gilbert, G.L. SARS, MERS and COVID-19—New Threats; Old Lessons. Int. J. Epidemiol. 2020, 49, 726–728.
- Kadam, S.B.; Sukhramani, G.S.; Bishnoi, P.; Pable, A.A.; Barvkar, V.T. SARS-CoV-2, the Pandemic Coronavirus: Molecular and Structural Insights. J. Basic Microbiol. 2021, 61, 180–202.
- Khailany, R.A.; Safdar, M.; Ozaslan, M. Genomic Characterization of a Novel SARS-CoV-2. Gene Rep. 2020, 19, 100682.
- Kim, D.; Lee, J.-Y.; Yang, J.-S.; Kim, J.W.; Kim, V.N.; Chang, H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020, 181, 914–921.e10.
- Bačenková, D.; Trebuňová, M.; Špakovská, T.; Schnitzer, M.; Bednarčíková, L.; Živčák, J. Comparison of Selected Characteristics of SARS-CoV-2, SARS-CoV, and HCoV-NL63. Appl. Sci. 2021, 11, 1497.
- Kirtipal, N.; Bharadwaj, S.; Kang, S.G. From SARS to SARS-CoV-2, Insights on Structure, Pathogenicity and Immunity Aspects of Pandemic Human Coronaviruses. Infect. Genet. Evol. 2020, 85, 104502.
- Greenhalgh, T.; Jimenez, J.L.; Prather, K.A.; Tufekci, Z.; Fisman, D.; Schooley, R. Ten Scientific Reasons in Support of Airborne Transmission of SARS-CoV-2. Lancet 2021, 397, 1603–1605.
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 Entry into Cells. Nat. Rev. Mol. Cell Biol. 2021, 23, 3–20.
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8.
- Papa, G.; Mallery, D.L.; Albecka, A.; Welch, L.G.; Cattin-Ortolá, J.; Luptak, J.; Paul, D.; McMahon, H.T.; Goodfellow, I.G.; Carter, A.; et al. Furin Cleavage of SARS-CoV-2 Spike Promotes but Is Not Essential for Infection and Cell-Cell Fusion. PLoS Pathog. 2021, 17, e1009246.
- Padmanabhan, P.; Desikan, R.; Dixit, N.M. Targeting TMPRSS2 and Cathepsin B/L Together May Be Synergistic against SARS-CoV-2 Infection. PLoS Comput. Biol. 2020, 16, e1008461.
- Prasad, K.; AlOmar, S.Y.; Almuqri, E.A.; Rudayni, H.A.; Kumar, V. Genomics-Guided Identification of Potential Modulators of SARS-CoV-2 Entry Proteases, TMPRSS2 and Cathepsins B/L. PLoS ONE 2021, 16, e0256141.
- Sungnak, W.; Huang, N.; Bécavin, C.; Berg, M.; Queen, R.; Litvinukova, M.; Talavera-López, C.; Maatz, H.; Reichart, D.; Sampaziotis, F.; et al. SARS-CoV-2 Entry Factors Are Highly Expressed in Nasal Epithelial Cells Together with Innate Immune Genes. Nat. Med. 2020, 26, 681–687.
- Lukassen, S.; Chua, R.L.; Trefzer, T.; Kahn, N.C.; Schneider, M.A.; Muley, T.; Winter, H.; Meister, M.; Veith, C.; Boots, A.W.; et al. SARS-CoV-2 Receptor ACE2 and TMPRSS2 Are Primarily Expressed in Bronchial Transient Secretory Cells. EMBO J. 2020, 39, e105114.
- Zhou, L.; Xu, Z.; Castiglione, G.M.; Soiberman, U.S.; Eberhart, C.G.; Duh, E.J. ACE2 and TMPRSS2 Are Expressed on the Human Ocular Surface, Suggesting Susceptibility to SARS-CoV-2 Infection. Ocul. Surf. 2020, 18, 537–544.
- Fodoulian, L.; Tuberosa, J.; Rossier, D.; Boillat, M.; Kan, C.; Pauli, V.; Egervari, K.; Lobrinus, J.A.; Landis, B.N.; Carleton, A.; et al. SARS-CoV-2 Receptors and Entry Genes Are Expressed in the Human Olfactory Neuroepithelium and Brain. iScience 2020, 23, 101839.
- World Health Organization. Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants (accessed on 23 May 2022).
- Khandia, R.; Singhal, S.; Alqahtani, T.; Kamal, M.A.; El-Shall, N.A.; Nainu, F.; Desingu, P.A.; Dhama, K. Emergence of SARS-CoV-2 Omicron (B.1.1.529) Variant, Salient Features, High Global Health Concerns and Strategies to Counter It amid Ongoing COVID-19 Pandemic. Environ. Res. 2022, 209, 112816.
- Choi, J.Y.; Smith, D.M. SARS-CoV-2 Variants of Concern. Yonsei Med. J. 2021, 62, 961–968.
- Johansson, M.A.; Quandelacy, T.M.; Kada, S.; Prasad, P.V.; Steele, M.; Brooks, J.T.; Slayton, R.B.; Biggerstaff, M.; Butler, J.C. SARS-CoV-2 Transmission from People Without COVID-19 Symptoms. JAMA Netw. Open 2021, 4, e2035057.
- Diamond, M.S.; Kanneganti, T.-D. Innate Immunity: The First Line of Defense against SARS-CoV-2. Nat. Immunol. 2022, 23, 165–176.
- Boechat, J.L.; Chora, I.; Morais, A.; Delgado, L. The Immune Response to SARS-CoV-2 and COVID-19 Immunopathology—Current Perspectives. Pulmonology 2021, 27, 423–437.
- Thorne, L.G.; Reuschl, A.-K.; Zuliani-Alvarez, L.; Whelan, M.V.X.; Turner, J.; Noursadeghi, M.; Jolly, C.; Towers, G.J. SARS-CoV-2 Sensing by RIG-I and MDA5 Links Epithelial Infection to Macrophage Inflammation. EMBO J. 2021, 40, e107826.
- Severa, M.; Diotti, R.A.; Etna, M.P.; Rizzo, F.; Fiore, S.; Ricci, D.; Iannetta, M.; Sinigaglia, A.; Lodi, A.; Mancini, N.; et al. Differential Plasmacytoid Dendritic Cell Phenotype and Type I Interferon Response in Asymptomatic and Severe COVID-19 Infection. PLOS Pathog. 2021, 17, e1009878.
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.-C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045.e9.
- Leisman, D.E.; Ronner, L.; Pinotti, R.; Taylor, M.D.; Sinha, P.; Calfee, C.S.; Hirayama, A.V.; Mastroiani, F.; Turtle, C.J.; Harhay, M.O.; et al. Cytokine Elevation in Severe and Critical COVID-19: A Rapid Systematic Review, Meta-Analysis, and Comparison with Other Inflammatory Syndromes. Lancet Respir. Med. 2020, 8, 1233–1244.
- García-Pérez, B.E.; González-Rojas, J.A.; Salazar, M.I.; Torres-Torres, C.; Castrejón-Jiménez, N.S. Taming the Autophagy as a Strategy for Treating COVID-19. Cells 2020, 9, 2679.
- Casadevall, A.; Joyner, M.J.; Pirofski, L.-A. SARS-CoV-2 Viral Load and Antibody Responses: The Case for Convalescent Plasma Therapy. J. Clin. Invest. 2020, 130, 5112–5114.
- Wang, H.; Yuan, Y.; Xiao, M.; Chen, L.; Zhao, Y.; Zhang, H.; Long, P.; Zhou, Y.; Xu, X.; Lei, Y.; et al. Dynamics of the SARS-CoV-2 Antibody Response up to 10 Months after Infection. Cell Mol. Immunol. 2021, 18, 1832–1834.
- Xu, L.; Li, D.; Ramadan, S.; Li, Y.; Klein, N. Facile Biosensors for Rapid Detection of COVID-19. Biosens. Bioelectron. 2020, 170, 112673.
- Yongchen, Z.; Shen, H.; Wang, X.; Shi, X.; Li, Y.; Yan, J.; Chen, Y.; Gu, B. Different Longitudinal Patterns of Nucleic Acid and Serology Testing Results Based on Disease Severity of COVID-19 Patients. Emerg. Microbes Infect. 2020, 9, 833–836.
- Herroelen, P.H.; Martens, G.A.; De Smet, D.; Swaerts, K.; Decavele, A.-S. Humoral Immune Response to SARS-CoV-2: Comparative Clinical Performance of Seven Commercial Serology Tests. Am. J. Clin. Pathol. 2020, 154, 610–619.
- Okba, N.M.A.; Müller, M.A.; Li, W.; Wang, C.; GeurtsvanKessel, C.H.; Corman, V.M.; Lamers, M.M.; Sikkema, R.S.; de Bruin, E.; Chandler, F.D.; et al. Severe Acute Respiratory Syndrome Coronavirus 2-Specific Antibody Responses in Coronavirus Disease Patients. Emerg. Infect. Dis. 2020, 26, 1478–1488.
- Long, Q.-X.; Liu, B.-Z.; Deng, H.-J.; Wu, G.-C.; Deng, K.; Chen, Y.-K.; Liao, P.; Qiu, J.-F.; Lin, Y.; Cai, X.-F.; et al. Antibody Responses to SARS-CoV-2 in Patients with COVID-19. Nat. Med. 2020, 26, 845–848.
- Loos, C.; Atyeo, C.; Fischinger, S.; Burke, J.; Slein, M.D.; Streeck, H.; Lauffenburger, D.; Ryan, E.T.; Charles, R.C.; Alter, G. Evolution of Early SARS-CoV-2 and Cross-Coronavirus Immunity. mSphere 2020, 5, e00622-20.
- Luo, H.; Jia, T.; Chen, J.; Zeng, S.; Qiu, Z.; Wu, S.; Li, X.; Lei, Y.; Wang, X.; Wu, W.; et al. The Characterization of Disease Severity Associated IgG Subclasses Response in COVID-19 Patients. Front. Immunol. 2021, 12, 632814.
- Kyei-Barffour, I.; Addo, S.A.; Aninagyei, E.; Ghartey-Kwansah, G.; Acheampong, D.O. Sterilizing Immunity against COVID-19: Developing Helper T Cells I and II Activating Vaccines Is Imperative. Biomed. Pharmacother. 2021, 144, 112282.
- Garcia-Beltran, W.F.; Lam, E.C.; St Denis, K.; Nitido, A.D.; Garcia, Z.H.; Hauser, B.M.; Feldman, J.; Pavlovic, M.N.; Gregory, D.J.; Poznansky, M.C.; et al. Multiple SARS-CoV-2 Variants Escape Neutralization by Vaccine-Induced Humoral Immunity. Cell 2021, 184, 2372–2383.e9.
- Planas, D.; Veyer, D.; Baidaliuk, A.; Staropoli, I.; Guivel-Benhassine, F.; Rajah, M.M.; Planchais, C.; Porrot, F.; Robillard, N.; Puech, J.; et al. Reduced Sensitivity of SARS-CoV-2 Variant Delta to Antibody Neutralization. Nature 2021, 596, 276–280.
- Sethuraman, N.; Jeremiah, S.S.; Ryo, A. Interpreting Diagnostic Tests for SARS-CoV-2. JAMA 2020, 323, 2249–2251.
- Caliendo, A.M.; Gilbert, D.N.; Ginocchio, C.C.; Hanson, K.E.; May, L.; Quinn, T.C.; Tenover, F.C.; Alland, D.; Blaschke, A.J.; Bonomo, R.A.; et al. Better Tests, Better Care: Improved Diagnostics for Infectious Diseases. Clin. Infect. Dis. 2013, 57, S139–S170.
- Kevadiya, B.D.; Machhi, J.; Herskovitz, J.; Oleynikov, M.D.; Blomberg, W.R.; Bajwa, N.; Soni, D.; Das, S.; Hasan, M.; Patel, M.; et al. Diagnostics for SARS-CoV-2 Infections. Nat. Mater. 2021, 20, 593–605.
- World Health Organization. Laboratory Testing for 2019 Novel Coronavirus (2019-NCoV) in Suspected Human Cases. Available online: https://www.who.int/publications-detail-redirect/10665-331501 (accessed on 23 May 2022).
- Ravi, N.; Cortade, D.L.; Ng, E.; Wang, S.X. Diagnostics for SARS-CoV-2 Detection: A Comprehensive Review of the FDA-EUA COVID-19 Testing Landscape. Biosens. Bioelectron. 2020, 165, 112454.
- Corman, V.M.; Drosten, C. Authors’ Response: SARS-CoV-2 Detection by Real-Time RT-PCR. Eurosurveillance 2020, 25, 2001035.
- Food and Drug Administration. In Vitro Diagnostics EUAs—Antigen Diagnostic Tests for SARS-CoV-2; FDA: Washington, DC, USA, 2022.
- CDC. Clinical Laboratory Improvement Amendments (CLIA). Available online: https://www.cdc.gov/clia/index.html (accessed on 27 August 2022).
This entry is offline, you can click here to edit this entry!
