Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Maternal-to-zygotic transition (MZT) of the control of early post-fertilization development is a key-event conditioning the fate of the future embryo, fetus and newborn. Because of the relative paucity of data concerning human embryos, due to ethical concerns and the poor availability of human embryos donated for research, most data have to be derived from animal models, among which those obtained using mouse embryos are most prevalent. Data obtained by studies performed in non-mammalian specie can also provide useful information.
- maternal-to-zygotic transition
- zygotic gene activation
- maternal mRNA decay
- M-decay
- Z-decay
- DNA transcription
- RNA translation
1. Introduction
Maternal-to-zygotic transition (MZT) refers to an initial step in the early embryonic development, consisting of two strictly coordinated phenomena, i.e., zygotic genome activation (ZGA) and the degradation of maternal gene transcripts. MZT is observed in all animal species investigated so far [1] including mammals [2]; it is essential for embryonic development because it coordinates cell division and allocation to the first two cell lineages, i.e., the inner cell mass (ICM) and trophectoderm, at the blastocyst stage [3]. Even though, so far as human embryos are concerned, there are still a lot of open questions, there is no doubt that failure of MZT is involved in preimplantation embryo demise and the resulting failure of assisted reproduction techniques (ART) [4]. Knowledge of the molecular mechanisms regulating MZT in humans is thus important for the development of diagnostic methods with which the quality of each individual embryo can be tested and the adequacy of particular ovarian stimulation protocols in each individual woman can be evaluated, enabling an individualized approach [5].
Because of the limited availability of human embryos to be used for scientific investigation, researchers have tried to extrapolate data obtained in various animal species to humans. However, it soon became evident that this approach was complicated, due to many significant interspecies differences in the mechanism and timing of MZT. Most studies were performed in different rodent species, mainly the mouse. However, the timing of human MZT is similar to that of some other mammals, including cow, sheep, rabbit, and macaque [6][7][8][9], suggesting that data obtained in these species may be more representative of humans [3].
Nevertheless, the mouse is the most available experimental animal model for the study of early embryonic development, and as such, data obtained with this model are important as a source of basic knowledge to be tested and appraised in a human clinical context, albeit while taking into account the interspecies differences [4]. Last but not least, a lot of studies have been published using non-mammalian animal species. Surprisingly, though phylogenetically more distant from humans, some molecular mechanisms of early embryonic development, including those related to MZT, are quite similar to those in humans; therefore, data obtained in those species should also be taken into account.
2. Similarities and Particularities of MZT in Human Preimplantation Embryos as Related to Other Animal Species
There are two classes of vertebrates: anamniotes and amniotes. The former lay oocytes externally in water and include bony fish and amphibians, whereas the latter lay fertilized oocytes on land or retain them in the mother in a protective membrane impermeable to water; these include mammals, birds and reptiles [3]. Anamniotic embryos contain all they need for the early development and typically show a period of rapid cell division following fertilization, whereas amniotic embryos develop more slowly [3]. In spite of this difference, some conserved principles of the vertebrate MZT appear to exist in both anamniotes and amniotes. Anamniotic embryos are also called fast-developing embryos, as opposed to the slow-developing amniotic embryos. In fast-developing embryos, early rapid synchronous cell divisions do not require embryonic gene transcription until gastrulation, and their cell cycles often lack G1 and G2 phases, alternating directly between the S and M phases [3].
A comparison of molecular events taking place during preimplnatation embryo development in human and mouse shows some remarkable differences (Figure 1), but also many similarities (Figure 2) which can serve to design future molecular studies aimed at developing new diagnostic and therapeutic strategies in the management of human infertility.
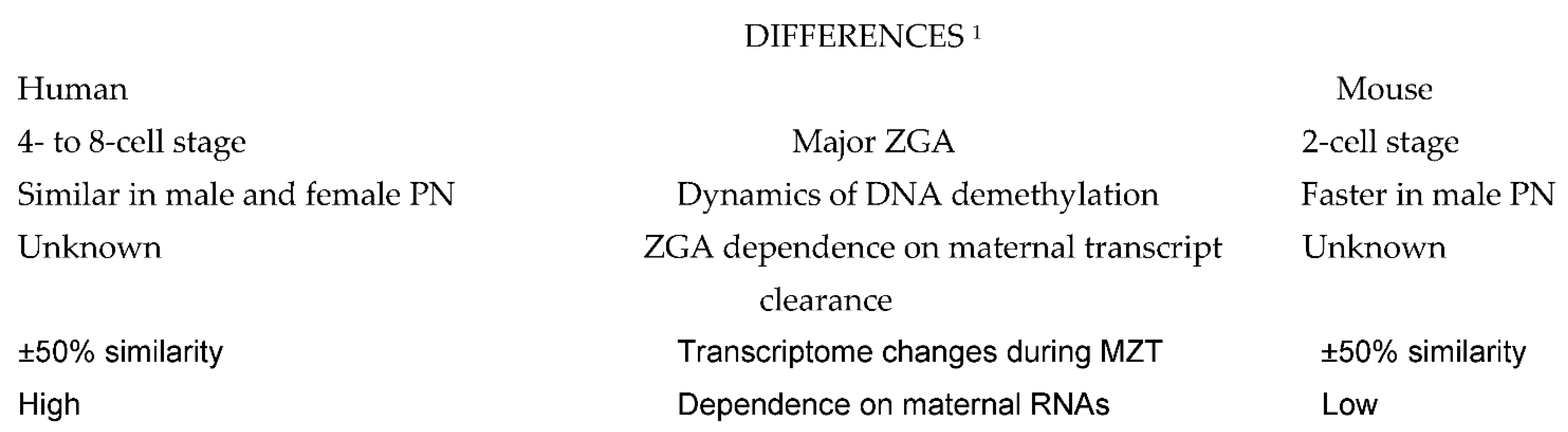
Figure 1. Differences between the basic molecular events taking place in human and mouse embryos. Explanations: 1 Comparisons are to be made taking into account the differences in ZGA timing. Abbreviations: ZGA: zygote gene activation; PN, pronuclei; MZT: maternal-to-zygotic transition of embryo developmental control.
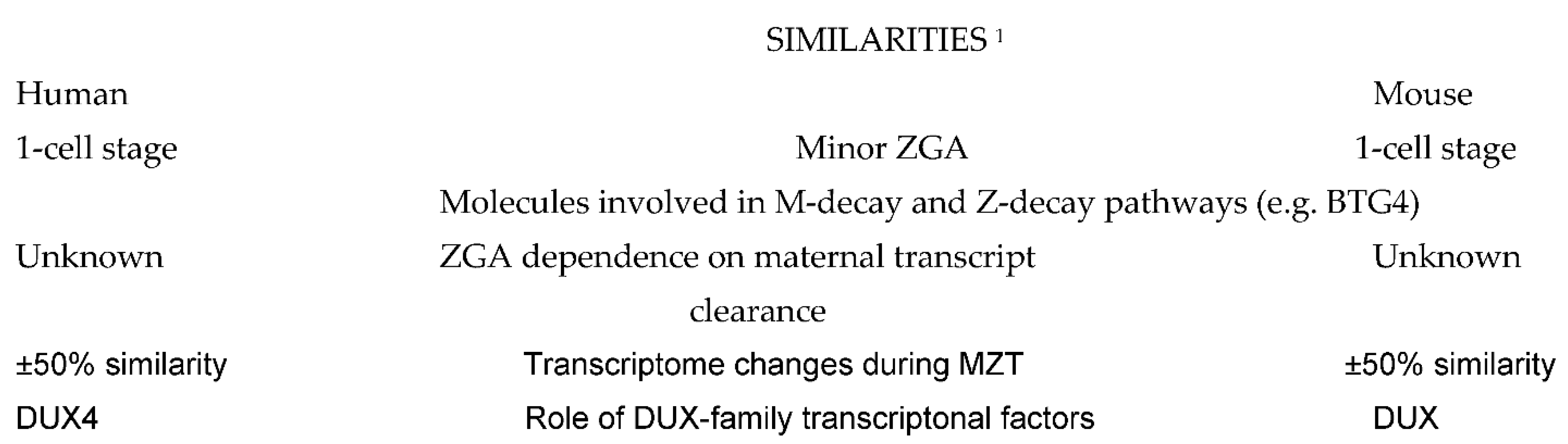
Figure 2. Similarities between the basic molecular events taking place in human and mouse embryos. Explanations: 1 Comparisons are to be made taking into account the differences in ZGA timing. Abbreviations: ZGA: zygote gene activation; MZT: maternal-to-zygotic transition of embryo developmental control; M-decay and Z-decay pathways: pathways of maternal transcript elimination controlled by maternal and zygotic transcripts, respectively. DUX: double homeobox.
In terms of generating a complete chain of gene transcription and expression, the beginning of ZGA in mammalian embryos occurs at the two-cell stage in mice [3], at the four-cell stage in pigs [10], at the eight-cell stage in sheep [6], cows [7] and rabbits [8], and between the four- cell and eight-cell stage in macaques [9] and humans [11][12][13]. In this sense, the early ZGA occurring in mice is rather exceptional, and its even later onset was also described in a variety of vertebrate anamniotic species (reviewed in [3]). However, the first detectable signs of gene transcription occur as early as the one-cell stage in many mammalian species, including mouse [14] and human [15], although most of these early transcripts are not translated into proteins, and their main function is to prepare embryos for the subsequent ZGA. In addition to participating in the nucleologenesis required for the production of the ribosomes needed during the forthcoming major wave of protein synthesis [15], this early wave of gene activity is also the source of other functional RNA coding for transcriptional factors participating both in the regulation of ZGA and maternal RNA degradation [16].
Before the onset of MZT, the early developmental processes in preimplantation embryos are entirely under the control of maternally inherited factors present in the mature oocyte. In fact, maternal RNAs and proteins exclusively guide the development while the zygotic genome remains quiescent [17]. Embryos of mammalian species that display early MZT onset, such as the mouse, are thus likely to be less dependent on maternal RNAs and proteins stored in the unfertilized oocyte as compared to species with a relatively late MZT onset, such as the human. This can explain why developmental competence has not yet been achieved with the use of in vitro culture systems to support preantral follicle development up to maturation in human and domestic animals [18]. The adequate progression of early post-fertilization developmental events occurring before the beginning of MZT is dependent on timely translation regulation of maternal mRNAs in time and space in all species studied so far [19]. The mechanisms involved in these regulatory processes, revealed in different species, involve mRNA association with, and its subsequent orchestrated release from, RNA-binding proteins [20], the spatial localization of the RNA-protein (RNP) complexes in the cytoplasm [21], and the timely activation of specific mRNA translation by modulation of cytoplasmic polyadenylation [22][23].
Both ZGA and maternal RNA clearance, the two basic events occurring during MZT, are highly complex processes that have to occur in a strictly orchestrated way in order to ensure normal embryonic development [24]. In Drosophila, maternal RNA clearance was shown to be a necessary pre-requisite for ZGA and MZT [25]. Studies on Drosophila [26] and mouse [27] embryos have shown that the elimination of maternal transcripts is accomplished by two sequential pathways. The first pathway, termed maternal decay (M-decay), is entirely mediated by maternal factors accumulated in the mature oocytes, whereas the second pathway, termed zygotic decay (Z-decay), depends on de novo zygotic transcription products appearing after fertilization. Key factors regulating M-decay and Z-decay pathways appear to have similar expression patterns in mouse and human MZT, involving YAP1-TEAD4 transcription activators, TUT4/7-mediated mRNA 3′-oligouridylation, and BTG4/CCR4-NOT-induced mRNA deadenylation [24]. The observation that homozygous mutations in human BTG4 cause zygotic cleavage arrest and female infertility [28] supports the hypothesis of functional equivalence of the pathways controlling M- and Z-decay in mice and humans. Both M- and Z-decay pathway activities appear to contribute to the developmental potential of human preimplantation embryos [24].
Despite extensive research on murine maternal and zygotic transcriptomes in recent years, it is still not clear to what extent the transcriptional pathways of mouse and human embryos are similar, and many questions regarding key MZT events in humans remain unanswered [29]. For instance, the proportion of human maternal transcripts with ZGA-dependent clearance remains undetermined [24]. In addition to similarities between the MZT regulating pathways in mice and humans, some differences are also likely to exist. The discovery of hominoid specific transposable elements and KZFPs that control human embryonic genome activation [30] points in this direction. Other differences between the early embryos of mice and humans concern the dynamics of the DNA methylation pattern, a crucial element in the epigenetic regulation of mammalian embryonic development. In fact, the major wave of genome-wide demethylation in human embryos is complete at the two-cell stage. Contrary to observations in mice, the demethylation of the paternal genome is much faster than that of the maternal genome, so the genome-wide methylation level in male pronuclei is already lower than that in female pronuclei [31].
Moreover, comparisons of human and mouse transcriptomes during MZT revealed that only half of the transcriptomes were overlapping, indicating that the homology between human and mouse maternal transcriptomes is low, and even fewer zygotically activated genes are shared by mouse two-cell embryos and human eight-cell embryos [24]. Also, some transcripts degraded by M-decay pathways in humans were found to be degraded by Z-decay pathways in mice, and vice versa, suggesting that subsets of human mRNA might be regulated differently from mouse during MZT [24]. In general, zygotic transcription plays a more important role in the elimination of human maternal transcripts, supposedly due to a longer time span from the onset of major ZGA to the completion of maternal mRNA decay in humans compared to mice [32].
As for the mechanisms regulating MZT timing, three models have been proposed. The first claims that MZT dynamics are triggered by the accumulation of maternally deposited activating transcription factors. The second model suggests that MZT dynamics are triggered by reaching a threshold ratio of a nuclear component to the cytoplasmic volume, often referred to as the “N/C-ratio”. The third model implies the de novo establishment of chromatin states which allow transcription to be critical for ZGA timing, because embryos must start from a state with no transcription. Importantly, these three timing mechanisms are not mutually exclusive and additional mechanisms may also exist [3].
Regarding mammalian embryos, maternally deposited activating transcription factors Nanog, SoxB1, and Oct4 (Pouf1) were suggested to activate transcription in preimplantation embryos of mice and human embryonic stem cells [33]. As for the “N/C-ratio” model, convincing data available for mammalian embryos are lacking. By contrast, the mechanisms implying the de novo establishment of chromatin states which are permissive for transcription are active in mammalian embryos, including humans (see below).
It is also important to note that the clearance timing differs among specific mRNA species, and there is a period of overlap during which both maternal and zygotic transcripts participate in the control of embryonic development. It was shown that in human embryos in which some blastomeres failed to undergo the major ZGA by the eight-cell stage, the development of intercellular junctions, required to seal the fluid-filled space in the future blastocyst, began in the same way in the cells that had accomplished ZGA and in those that had not [34]. However, this process was incomplete in ZGA-failed blastomeres. Thus, oocyte-coded messages are apparently involved in the control of the relatively late stages of human preimplantation development, including the differentiation of the first two embryonic cell lines, while the embryonic genome is required for the full achievement of this early differentiation event. In any case, a temporal overlap in the translation of maternal and zygotic transcripts is a common feature of embryos of different animal species [35].
The role of the double homeobox (DUX) family of transcription factors is also highly conserved among different mammalian species, including mouse and primates [36], where they act as pioneer actors of ZGA [37][38]. Mouse DUX and its human homolog DUX4 activate ZGA by acting as regulators of the non-coding genome [39].
Taken together, studies performed so far have revealed both similarities and differences in the control of MZT among mammalian species. Concerning possible future implications in human infertility management and research, the use of data obtained in mice, by far the most studied mammalian species, will certainly serve as the main basis.
This entry is adapted from the peer-reviewed paper 10.3390/ijms23158562
References
- Tadros, W.; Lipshitz, H.D. The maternal-to-zygotic transition: A play in two acts. Development 2009, 136, 3033–304242.
- Sha, Q.Q.; Zhang, J.; Fan, H.Y. A story of birth and death: mRNA translation and clearance at the onset of maternal-to-zygotic transition in mammals. Biol. Reprod. 2019, 101, 579–590.
- Jukam, D.; Shariati, S.A.M.; Skotheim, J.M. Zygotic Genome Activation in Vertebrates. Dev. Cell 2017, 42, 316–332.
- Paonessa, M.; Borini, A.; Coticchio, G. Genetic causes of preimplantation embryo developmental failure. Mol. Reprod. Dev. 2021, 88, 338–348.
- Tesarik, J. Toward molecular medicine in female infertility management: Editorial to the Special Issue “Molecular Mechanisms of Human Oogenesis and Early Embryogenesis”. Int. J. Mol. Sci. 2021, 22, 13517.
- Crosby, I.M.; Gandolfi, F.; Moor, R.M. Control of protein synthesis during early cleavage of sheep embryos. J. Reprod. Fertil. 1988, 82, 769–775.
- Frei, R.E.; Schultz, G.A.; Church, R.B. Qualitative and quantitative changes in protein synthesis occur at the 8–16-cell stage of embryogenesis in the cow. J. Reprod. Fertil. 1989, 86, 637–641.
- Christians, E.; Rao, V.H.; Renard, J.P. Sequential acquisition of transcriptional control during early embryonic development in the rabbit. Dev. Biol. 1994, 164, 160–172.
- Schramm, R.D.; Bavister, B.D. Onset of nucleolar and extranucleolar transcription and expression of fibrillarin in macaque embryos developing in vitro. Biol. Reprod. 1999, 60, 721–728.
- Cao, S.; Han, J.; Wu, J.; Li, Q.; Liu, S.; Zhang, W.; Pei, Y.; Ruan, X.; Liu, Z.; Wang, X.; et al. Specific gene-regulation networks during the pre-implantation development of the pig embryo as revealed by deep sequencing. BMC Genom. 2014, 15, 4.
- Tesarik, J.; Kopecny, V.; Plachot, M.; Mandelbaum, J. Activation of nucleolar and extranucleolar RNA synthesis and changes in the ribosomal content of human embryos developing in vitro. J. Reprod. Fertil. 1986, 78, 463–470.
- Braude, P.; Bolton, V.; Moore, S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature 1988, 332, 459–461.
- Tesarik, J.; Kopecny, V.; Plachot, M.; Mandelbaum, J. Early morphological signs of embryonic genome expression in human preimplantation development as revealed by quantitative electron microscopy. Dev. Biol. 1988, 128, 15–20.
- Abe, K.I.; Funaya, S.; Tsukioka, D.; Kawamura, M.; Suzuki, Y.; Suzuki, M.G.; Schultz, R.M.; Aoki, F. Minor zygotic gene activation is essential for mouse preimplantation development. Proc. Natl. Acad. Sci. USA 2018, 115, E6780–E6788.
- Tesarik, J.; Kopecny, V. Assembly of the nucleolar precursor bodies in human male pronuclei is correlated with an early RNA synthetic activity. Exp. Cell Res. 1990, 191, 153–156.
- Asami, M.; Lam, B.Y.H.; Ma, M.K.; Rainbow, K.; Braun, S.; VerMilyea, M.D.; Yeo, G.S.H.; Perry, A.C.F. Human embryonic genome activation initiates at the one-cell stage. Cell Stem Cell 2022, 29, 209–216.e4.
- Shi, Y.; Cai, M.; Du, K.; Bai, X.; Tang, L.; Jia, X.; Chen, S.; Wang, J.; Lai, S. Dynamics of known long non-coding RNAs during the maternal-to-zygotic transition in rabbit. Animals 2021, 11, 3592.
- Paulino, L.R.F.M.; de Assis, E.I.T.; Azevedo, V.A.N.; Silva, B.R.; da Cunha, E.V.; Silva, J.R.V. Why is it so difficult to have competent oocytes from in vitro cultured preantral follicles? Reprod. Sci. 2022. Epub ahead of print.
- Winata, C.L.; Korzh, V. The translational regulation of maternal mRNAs in time and space. FEBS Lett. 2018, 592, 3007–3023.
- Shav-Tal, Y.; Singer, R.H. RNA localization. J. Cell Sci. 2005, 118, 4077–4081.
- Anderson, P.; Kedersha, N. RNA granules: Post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 2009, 10, 430–436.
- Lieberfarb, M.E.; Chu, T.; Wreden, C.; Theurkauf, W.; Gergen, J.P.; Strickland, S. Mutations that perturb poly(A)-dependent maternal mRNA activation block the initiation of development. Development 1996, 122, 579–588.
- Aoki, F.; Hara, K.T.; Schultz, R.M. Acquisition of transcriptional competence in the 1-cell mouse embryo: Requirement for recruitment of maternal mRNAs. Mol. Reprod. Dev. 2003, 64, 270–274.
- Sha, Q.Q.; Zheng, W.; Wu, Y.W.; Li, S.; Guo, L.; Zhang, S.; Lin, G.; Ou, X.H.; Fan, H.Y. Dynamics and clinical relevance of maternal mRNA clearance during the oocyte-to-embryo transition in humans. Nat. Commun. 2020, 11, 4917.
- Blatt, P.; Wong-Deyrup, S.W.; McCarthy, A.; Breznak, S.; Hurton, M.D.; Upadhyay, M.; Bennink, B.; Camacho, J.; Lee, M.T.; Rangan, P. RNA degradation is required for the germ-cell to maternal transition in Drosophila. Curr. Biol. 2021, 31, 2984–2994.e7.
- Bashirullah, A.; Halsell, S.R.; Cooperstock, R.L.; Kloc, M.; Karaiskakis, A.; Fisher, W.W.; Fu, W.; Hamilton, J.K.; Etkin, L.D.; Lipshitz, H.D. Joint action of two RNA degradation pathways controls the timing of maternal transcript elimination at the midblastula transition in Drosophila melanogaster. EMBO J. 1999, 18, 2610–2620.
- Alizadeh, Z.; Kageyama, S.; Aoki, F. Degradation of maternal mRNA in mouse embryos: Selective degradation of specific mRNAs after fertilization. Mol. Reprod. Dev. 2005, 72, 281–290.
- Zheng, W.; Zhou, Z.; Sha, Q.; Niu, X.; Sun, X.; Shi, J.; Zhao, L.; Zhang, S.; Dai, J.; Cai, S.; et al. Homozygous mutations in BTG4 cause zygotic cleavage failure and female infertility. Am. J. Hum. Genet. 2020, 107, 24–33.
- Yan, L.; Yang, M.; Guo, H.; Yang, L.; Wu, J.; Li, R.; Liu, P.; Lian, Y.; Zheng, X.; Yan, J.; et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 2013, 20, 1131–1139.
- Pontis, J.; Planet, E.; Offner, S.; Turelli, P.; Duc, J.; Coudray, A.; Theunissen, T.W.; Jaenisch, R.; Trono, D. Hominoid-Specific Transposable Elements and KZFPs Facilitate Human Embryonic Genome Activation and Control Transcription in Naive Human ESCs. Cell Stem Cell 2019, 24, 724–735.e5.
- Guo, H.; Zhu, P.; Yan, L.; Li, R.; Hu, B.; Lian, Y.; Yan, J.; Ren, X.; Lin, S.; Li, J.; et al. The DNA methylation landscape of human early embryos. Nature 2014, 511, 606–610.
- Zhang, Y.; Yan, Z.; Qin, Q.; Nisenblat, V.; Chang, H.M.; Yu, Y.; Wang, T.; Lu, C.; Yang, M.; Yang, S.; et al. Transcriptome landscape of human folliculogenesis reveals oocyte and granulosa cell interactions. Mol. Cell 2018, 72, 1021–1034.e4.
- Boyer, L.A.; Lee, T.I.; Cole, M.F.; Johnstone, S.E.; Levine, S.S.; Zucker, J.P.; Guenther, M.G.; Kumar, R.M.; Murray, H.L.; Jenner, R.G.; et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 2005, 122, 947–956.
- Tesarík, J. Involvement of oocyte-coded message in cell differentiation control of early human embryos. Development 1989, 105, 317–322.
- Lee, M.T.; Bonneau, A.R.; Giraldez, A.J. Zygotic genome activation during the maternal-to-zygotic transition. Annu. Rev. Cell Dev. Biol. 2014, 30, 581–613.
- Whiddon, J.L.; Langford, A.T.; Wong, C.J.; Zhong, J.W.; Tapscott, S.J. Conservation and innovation in the DUX4-family gene network. Nat. Genet. 2017, 49, 935–940.
- De Iaco, A.; Planet, E.; Coluccio, A.; Verp, S.; Duc, J.; Trono, D. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet. 2017, 49, 941–945.
- Hendrickson, P.G.; Doráis, J.A.; Grow, E.J.; Whiddon, J.L.; Lim, J.W.; Wike, C.L.; Weaver, B.D.; Pflueger, C.; Emery, B.R.; Wilcox, A.L.; et al. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 2017, 49, 925–934.
- Vuoristo, S.; Bhagat, S.; Hydén-Granskog, C.; Yoshihara, M.; Gawriyski, L.; Jouhilahti, E.M.; Ranga, V.; Tamirat, M.; Huhtala, M.; Kirjanov, I.; et al. DUX4 is a multifunctional factor priming human embryonic genome activation. iScience 2022, 25, 104137.
This entry is offline, you can click here to edit this entry!
