Phytoplasmas are pleomorphic, wall-less intracellular bacteria that can cause devastating diseases in a wide variety of plant species. Rapid diagnosis and precise identification of phytoplasmas responsible for emerging plant diseases are crucial to preventing further spread of the diseases and reducing economic losses. Phytoplasma taxonomy (identification, nomenclature, and classification) has lagged in comparison to culturable bacteria, largely due to lack of axenic phytoplasma culture and consequent inaccessibility of phenotypic characteristics. However, the rapid expansion of molecular techniques and the advent of high throughput genome sequencing have tremendously enhanced the nucleotide sequence-based phytoplasma taxonomy.
- phytoplasma
- bacterial taxonomy
- whole genome-based average nucleotide identity (ANI)
- iPhyClassifier
1. Phytoplasma Nomenclature: Delineation of Candidatus Phytoplasma Species
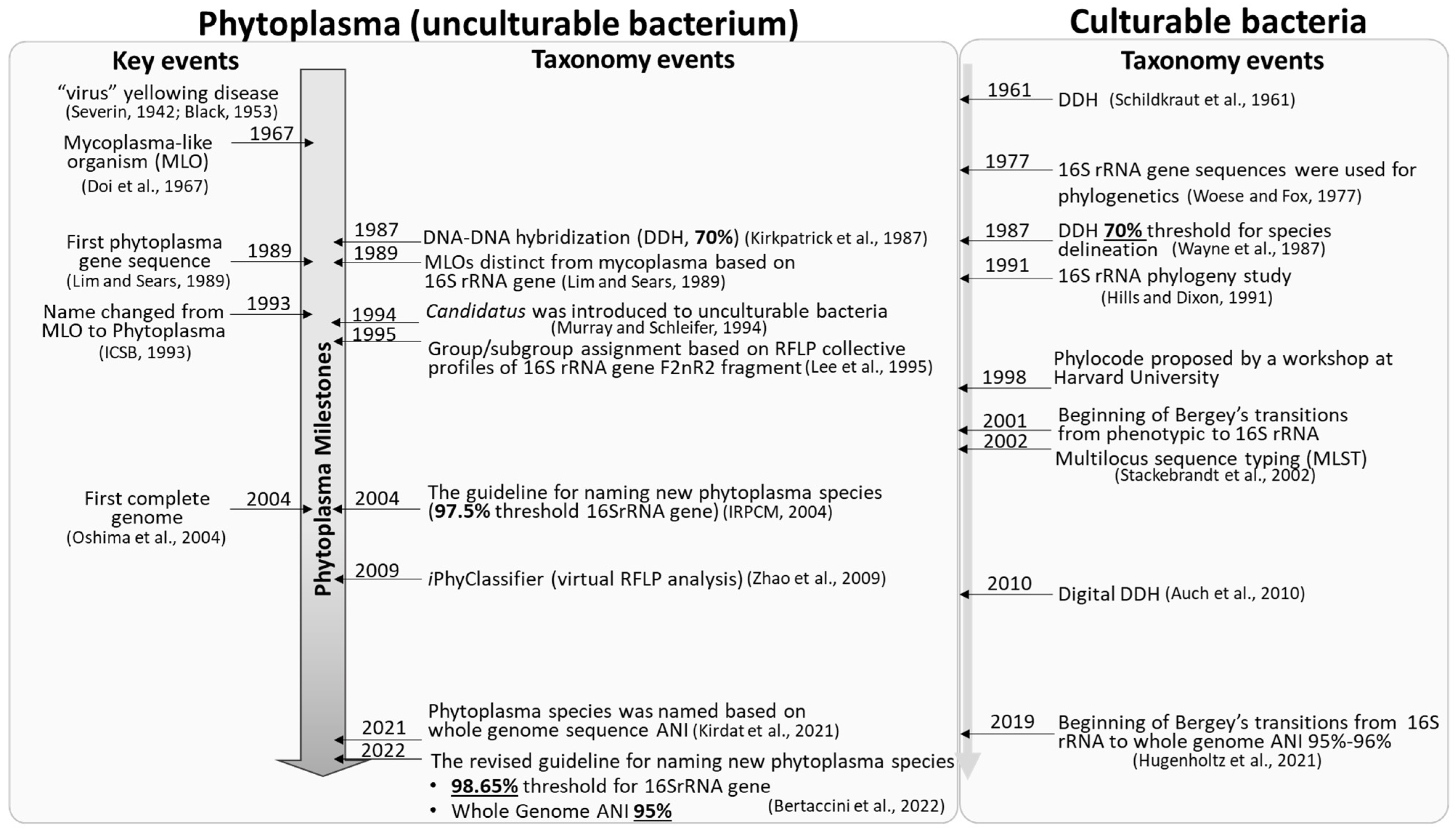
2. Phytoplasma Classification: 16Sr Group/Subgroup Classification System Based on Collective RFLP Profiles
| Group | Number of ‘Ca. Phytoplasma’ Species | Accession Number of Reference Strain | Subgroup | Reference | |
|---|---|---|---|---|---|
| 16SrI: Aster yellows group | 3 | ‘Ca. Phytoplasma asteris’ | M30790 | 16SI-B | [36] |
| ‘Ca. Phytoplasma lycopersici’ | EF199549 | 16SrI-Y | [37] | ||
| ‘Ca. Phytoplasma tritici’ | NZ AVAO01000003 | 16SrI-C | [75] | ||
| 16SrII: Peanut witches’ broom group | 1 | ‘Ca. Phytoplasma aurantifolia’ | U15442 | 16SrII-B | [76] |
| * Abolished | ‘Ca. Phytoplasma australasia’ | Y10096 | 16SrII-D | [38] | |
| 16SrIII: X-disease group | 1 | ‘Ca. Phytoplasma pruni’ | JQ044393 | 16SrIII-A | [39] |
| 16SrIV: Coconut lethal yellows group | 2 | ‘Ca. Phytoplasma palmae’ | U18747 | 16SrIV-A | [18][77] |
| ‘Ca. Phytoplasma cocostanzaniae’ | X80117 | 16SrIV-C | [18][77] | ||
| 16SrV: Elm yellows group | 4 | ‘Ca. Phytoplasma ulmi’ | AY197655 | 16SrV-A | [40] |
| ‘Ca. Phytoplasma ziziphi’ | AB052876 | 16SrV-B | [41] | ||
| ‘Ca. Phytoplasma rubi’ | AY197648 | 16SrV-E | [42] | ||
| ‘Ca. Phytoplasma balanitae’ | AB689678 | 16SrV-new subgroup | [43] | ||
| 16SrVI: Clover proliferation group | 2 | ‘Ca. Phytoplasma trifolii’ | AY390261 | 16SrVI-A | [44] |
| ‘Ca. Phytoplasma sudamericanum’ | GU292081 | 16SrVI-I | [45] | ||
| 16SrVII: Ash yellows group | 1 | ‘Ca. Phytoplasma fraxini’ | AF092209 | 16SrVII-A | [46] |
| 16SrVIII: Loofah witches’ broom group | 1 | ‘Ca. Phytoplasma luffae’ | AF248956 | 16SrVIII-A | [47] |
| 16SrIX: Pigeon pea witches’ broom group | 1 | ‘Ca. Phytoplasma phoenicium’ | AF248956 | 16SrIX-D | [48] |
| 16SrX: Apple proliferation group | 4 | ‘Ca. Phytoplasma mali’ | AJ542541 | 16SrX-A | [49] |
| ‘Ca. Phytoplasma pyri’ | AJ542543 | 16SrX-C | [49] | ||
| ‘Ca. Phytoplasma prunorum’ | AJ542544 | 16SrX-F | [49] | ||
| ‘Ca. Phytoplasma spartii’ | X92869 | 16SrX-D | [50] | ||
| 16SrXI: Rice yellow dwarf group | 3 | ‘Ca. Phytoplasma oryzae’ | AB052873 | 16SrXI-A | [51] |
| ‘Ca. Phytoplasma cirsii’ | KR869146 | 16SrXI-D | [52] | ||
| ‘Ca. Phytoplasma sacchari’ | VWXM00000000 | 16SrXI-B | [78] | ||
| 16SrXII: Stolbur group | 5 | ‘Ca. Phytoplasma australiense’ | L76865 | 16SrXII-B | [53] |
| ‘Ca. Phytoplasma japonicum’ | AB010425 | 16SrXII-D | [54] | ||
| ‘Ca. Phytoplasma fragariae’ | DQ086423 | 16SrXII-E | [55] | ||
| ‘Ca. Phytoplasma solani’ | AF248959 | 16SrXII-A | [56] | ||
| ‘Ca. Phytoplasma convolvuli’ | JN833705 | 16SrXII-H | [57] | ||
| 16SrXIII: Mexican periwinkle virescence group | 2 | ‘Ca. Phytoplasma hispanicum’ | AF248960 | 16SrXIII-A | [58] |
| ‘Ca. Phytcoplasma meliae | KU850940 | 16SrXIII-G | [59] | ||
| 16SrXIV: Bermudagrass white leaf group | 1 | ‘Ca. Phytoplasma cynodontis’ | AJ550984 | 16SrXIV-A | [60] |
| 16SrXV: Hibiscus witches’ broom group | 1 | ‘Ca. Phytoplasma brasiliense’ | AF147708 | 16SrXV-A | [61] |
| 16SrXVI: Sugar cane yellow leaf syndrome group | 1 | ‘Ca. Phytoplasma graminis’ | AY725228 | 16SrXVI-A | [62] |
| 16SrXVII: Papaya bunchy top group | 1 | ‘Ca. Phytoplasma caricae’ | AY725234 | 16SrXVII-A | [62] |
| 16SrXVIII: American potato purple top wilt group | 1 | ‘Ca. Phytoplasma americanum’ | DQ174122 | 16SrXVIII-A | [63] |
| 16SrXIX: Japanese chestnut witches’ broom group | 1 | ‘Ca. Phytoplasma castaneae’ | AB054986 | 16SrXIX-A | [64] |
| 16SrXX: Buckthorn witches’ broom group | 1 | ‘Ca. Phytoplasma rhamni’ | X76431 | 16SrXX-A | [50] |
| 16SrXXI: Pine shoot proliferation group | 1 | ‘Ca. Phytoplasma pini’ | AJ632155 | 16SrXXI-A | [65] |
| 16SrXXII: Nigerian coconut lethal decline group | 1 | ‘Ca. Phytoplasma palmicola’ | KF751387 | 16SrXXII-A | [66] |
| 16SrXXIII: Buckland Valley grapevine yellows group | 1 unnamed species identified | AY083605 | 16SrXXIII-A | [31] | |
| 16SrXXIV: Sorghum bunchy shoot group | 1 unnamed new species identified | AF509322 | 16SrXXIV-A | ||
| 16SrXXV: Weeping tea tree witches’ broom group | 1 unnamed new species identified | AF521672 | 16SrXXV-A | ||
| 16SrXXVI: Mauritius sugar cane yellows D3T1 group | 1 unnamed new species identified | AJ539179 | 16SrXXVI-A | ||
| 16SrXXVII: Mauritius sugar cane yellows D3T2 group | 1 unnamed new species identified | AJ539180 | 16SrXXVII-A | ||
| 16SrXXVIII: Havana derbid group | 1 unnamed new species identified | AY744945 | 16SrXXVII-A | ||
| 16SrXXIX: Cassia witches’ broom group | 1 | ‘Ca. Phytoplasma omanense’ | EF666051 | 16SrXXIX-A | [67] |
| 16SrXXX: Salt cedar witches’ broom group | 1 | ‘Ca. Phytoplasma tamaricis’ | FJ432664 | 16SrXXX-A | [68] |
| 16SrXXXI: Soybean stunt phytoplasma group | 1 | ‘Ca. Phytoplasma costaricanum’ | HQ225630 | 16SrXXXI-A | [69] |
| 16SrXXXII: Malaysian periwinkle virescence group | 1 | ‘Ca. Phytoplasma malaysianum’ | EU371934 | 16SrXXXII-A | [70] |
| 16SrXXXIII: Allocasuarina group | 1 | ‘Ca. Phytoplasma allocasuarinae’ | AY135523 | 16SrXXXIII-A | [18] |
| 16SrXXXIV: grapevine yellows | No new species identified, abolished | DQ232752 | |||
| 16SrXXXV: Pepper witches’-broom | No new species identified, abolished | EU125184 | |||
| 16SrXXXVI: foxtail palm yellow decline group | 1 | ‘Ca. Phytoplasma wodyetiae’ | KC844879 | 16SrXXXVI-A | [71] |
| 16SrXXXVII: Stylosanthes little leaf group | 1 | ‘Ca. Phytoplasma stylosanthis’ | MT431550 | 16SrXXXVII-A | [72] |
| 16SrXXXVIII: Bogia coconut syndrome group | 1 | ‘Ca. Phytoplasma noviguineense’ | LC228755 | 16SrXXXVIII-A | [73] |
| 16SrXXXIX: Palm lethal wilt group | 1 | ‘Ca. Phytoplasma dypsidis’ | MT536195 | 16SrXXXIX-A | [74] |
3. Phytoplasma Identification: Detection, Diagnostics and Characterization
This entry is adapted from the peer-reviewed paper 10.3390/biology11081119
References
- Kämpfer, P.; Glaeser, S.P. Prokaryotic taxonomy in the sequencing era–the polyphasic approach revisited. Environ. Microbiol. 2012, 14, 291–317.
- Rosselló-Mora, R.; Amann, R. The species concept for prokaryotes. FEMS Microbiol. Rev. 2001, 25, 39–67.
- Chiykowski, L.N. Clover phyllody virus in Canada and its transmission. Can. J. Bot. 1962, 40, 397–404.
- Freitag, J.H. Interaction and mutual suppression among three strains of aster yellows virus. Virology 1964, 24, 401–413.
- Granados, R.R.; Chapman, R.K. Identification of some new aster yellows virus strains and their transmission by aster leafhopper Macrosteles fascifrons. Phytopathology 1968, 58, 1685.
- Chiykowski, L.N.; Sinha, R.C. Differentiation of MLO diseases by means of symptomatology and vector transmission. In Recent advances in mycoplasmology. In Proceedings of the 7th congress of the International Organization for Mycoplasmology, Baden near Vienna, Austria, 2–9 June 1988; Gustav Fischer Verlag: Baden near Vienna, Austria, 1990; pp. 280–287.
- McCoy, R.E.; Caudwell, A.; Chang, C.G.; Chen, T.A.; Chiykowski, L.N.; Cousin, M.T.; De Leeuw, G.D.; Golino, D.A.; Hacke, K.J.; Kirkpatrick, B.C.; et al. Mycoplasmalike organisms. Mycoplasmas 1989, 5, 545–568.
- Stackebrandt, E.; GOEBEL, B.M. Taxonomic note: A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 1994, 44, 846–849.
- Stephen, J.R.; McCaig, A.E.; Smith, Z.; Prosser, J.I.; Embley, T.M. Molecular diversity of soil and marine 16S rRNA gene sequences related to beta-subgroup ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 1996, 62, 4147–4154.
- Drancourt, M.; Bollet, C.; Carlioz, A.; Martelin, R.; Gayral, J.P.; Raoult, D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J. Clin. Microbiol. 2000, 38, 3623–3630.
- Moore, E.R.; Krüger, A.S.; Hauben, L.; Seal, S.E.; De Baere, R.; De Wachter, R.; Timmis, K.N.; Swings, J. 16S rRNA gene sequence analyses and inter-and intrageneric relationships of Xanthomonas species and Stenotrophomonas maltophilia. FEMS Microbiol. Lett. 1997, 151, 145–153.
- Woo, P.C.; Lau, S.K.; Teng, J.L.; Tse, H.; Yuen, K.Y. Then and now: Use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infect. 2008, 14, 908–934.
- Oren, A.; Garrity, G.M. Valid publication of the names of forty-two phyla of prokaryotes. Int. J. Syst. Evol. Microbiol. 2021, 71, 005056.
- Murray, R.G.E.; Sneath, P.H.A.; Mair, N.S.; Sharpe, M.E. Kingdom Procaryotae. Bergey’s Man. Syst. Bacteriol. 1984, 1, 34–36.
- Gasparich, G.E.; Bertaccini, A.; Zhao, Y. Candidatus Phytoplasma. In Bergey’s Manual of Systematics of Archaea and Bacteria; Trujillo, M.E., Dedysh, S., DeVos, P., Hedlund, B., Kämpfer, P., Rainey, F.A., Whitman, W.B., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2020.
- Murray, R.G.E.; Schleifer, K.H. Taxonomic notes: A proposal for recording the properties of putative taxa of procaryotes. Int. J. Syst. Evol. Microbiol. 1994, 44, 174–176.
- Oren, A. A plea for linguistic accuracy–also for Candidatus taxa. Int. J. Syst. Evol. Microbiol. 2017, 67, 1085–1094.
- IRPCM Phytoplasma/Spiroplasma Working Team–Phytoplasma Taxonomy Group. ‘Candidatus Phytoplasma’, a taxon for the wall-less, non-helical prokaryotes that colonize plant phloem and insects. Int. J. Syst. Evol. Microbiol. 2004, 54, 1243–1255.
- Baron, E.J. Classification. In Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996; Chapter 3. Available online: https://www.ncbi.nlm.nih.gov/books/NBK8406/ (accessed on 6 June 2022).
- Kirkpatrick, B.C. Strategies for characterizing plant pathogenic mycoplasma-like organisms and their effects on plants. Plant-Microbe Interact. USA 1989.
- Davis, R.E.; Lee, I.; Dally, E.L.; Dewitt, N.; Douglas, S.M. Cloned nucleic acid hybridization probes in detection and classification of mycoplasmalike organisms (MLOs). In Proceedings of the VII International Symposium on Virus Diseases of Ornamental Plants, Sanremo, Italy, 29 May–2 June 1988; Volume 234, pp. 115–122.
- Zhao, Y.; Wei, W.; Davis, R.E.; Lee, I.-M. Recent advances in 16S rRNA gene-based phytoplasma differ-entiation, classification and taxonomy. In Phytoplasmas: Genomes, Plant Hosts and Vector; Weintraub, P., Jones, P., Eds.; CABI Publishing: Wallingford, UK, 2010; pp. 64–92.
- Lee, I.M.; Bertaccini, A.; Vibio, M.; Gundersen, D.E. Detection of multiple phytoplasmas in perennial fruit trees with decline symptoms in Italy. Phytopathology 1995, 85, 728–735.
- Lee, I.M.; Gundersen-Rindal, D.E.; Davis, R.E.; Bartoszyk, I.M. Revised classification scheme of phytoplasmas based on RFLP analyses of 16S rRNA and ribosomal protein gene sequences. Int. J. Syst. Evol. Microbiol. 2010, 48, 1153–1169.
- Marcone, C.; Ragozzino, A.; Seemüller, E. Detection of Bermuda grass white leaf disease in Italy and characterization of the associated phytoplasma by RFLP analysis. Plant Dis. 1997, 81, 862–866.
- Pérez-López, E.; Luna-Rodríguez, M.; Olivier, C.Y.; Dumonceaux, T.J. The underestimated diversity of phytoplasmas in Latin America. Int. J. Syst. Evol. Microbiol. 2016, 66, 492–513.
- Fernandez, F.D.; Meneguzzi, N.G.; Guzman, F.A.; Kirschbaum, D.S.; Conci, V.C.; Nome, C.F.; Conci, L.R. Detection and identification of a novel 16SrXIII subgroup phytoplasma associated with strawberry red leaf disease in Argentina. Int. J. Syst. Evol. Microbiol. 2015, 65 Pt 8, 2741–2747.
- Nejat, N.; Vadamalai, G. Diagnostic techniques for detection of phytoplasma diseases: Past and present. J. Plant Dis. Prot. 2013, 120, 16–25.
- Lee, I.-M.; Davis, R.E.; Gundersen-Rindal, D.E. Phytoplasma: Phytopathogenic mollicutes. Annu. Rev. Microbiol. 2000, 54, 221–255.
- Zhao, Y.; Wei, W.; Lee, M.; Shao, J.; Suo, X.; Davis, R.E. Construction of an interactive online phytoplasma classification tool, iPhyClassifier, and its application in analysis of the peach X-disease phytoplasma group (16SrIII). Int. J. Syst. Evol. Microbiol. 2009, 59 Pt 10, 2582.
- Wei, W.; Davis, R.E.; Lee, M.; Zhao, Y. Computer-simulated RFLP analysis of 16S rRNA genes: Identification of ten new phytoplasma groups. Int. J. Syst. Evol. Microbiol. 2007, 57, 1855–1867.
- Wei, W.; Lee, M.; Davis, R.E.; Suo, X.; Zhao, Y. Automated RFLP pattern comparison and similarity coefficient calculation for rapid delineation of new and distinct phytoplasma 16Sr subgroup lineages. Int. J. Syst. Evol. Microbiol. 2008, 58, 2368–2377.
- Edel, V. Use of PCR and RFLP in fungal systematics. In Chemical Fungal Taxonomy; CRC Press: Boca Raton, FL, USA, 2020; pp. 51–76.
- Claussen, M.; Schmidt, S. Differentiation of Basidiobolus s isolates: RFLP of a diagnostic PCR amplicon matches sequence-based classification and growth temperature preferences. J. Fungi 2021, 7, 110.
- Silvester, R.; Alexander, D.; Antony, A.C.; Hatha, M. GroEL PCR-RFLP–an efficient tool to discriminate closely related pathogenic Vibrio species. Microb. Pathog. 2017, 105, 196–200.
- Lee, I.M.; Gundersen-Rindal, D.E.; Davis, R.E.; Bottner, K.D.; Marcone, C.; Seemüller, E. ‘Candidatus Phytoplasma asteris’, a novel phytoplasma taxon associated with aster yellows and related diseases. Int. J. Syst. Evol. Microbiol. 2004, 54, 1037–1048.
- Arocha, Y.; Antesana, O.; Montellano, E.; Franco, P.; Plata, G.; Jones, P. ‘Candidatus Phytoplasma lycopersici’, a phytoplasma associated with ‘hoja de perejil’ disease in Bolivia. Int. J. Syst. Evol. Microbiol. 2007, 57, 1704–1710.
- White, D.T.; Blackall, L.L.; Scott, P.T.; Walsh, K.B. Phylogenetic positions of phytoplasmas associated with dieback, yellow crinkle and mosaic diseases of papaya, and their proposed inclusion in ‘Candidatus Phytoplasma australiense’ and a new taxon, ‘Candidatus Phytoplasma australasia’. Int. J. Syst. Evol. Microbiol. 1998, 48, 941–951.
- Davis, R.E.; Zhao, Y.; Dally, E.L.; Lee, M.; Jomantiene, R.; Douglas, S.M. ‘Candidatus Phytoplasma pruni’, a novel taxon associated with X-disease of stone fruits, Prunus s: Multilocus characterization based on 16S rRNA, secY, and ribosomal protein genes. Int. J. Syst. Evol. Microbiol. 2013, 63 Pt 2, 766–776.
- Lee, M.; Martini, M.; Marcone, C.; Zhu, S.F. Classification of phytoplasma strains in the elm yellows group (16SrV) and proposal of ‘Candidatus Phytoplasma ulmi’ for the phytoplasma associated with elm yellows. Int. J. Syst. Evol. Microbiol. 2004, 54, 337–347.
- Jung, H.Y.; Sawayanagi, T.; Kakizawa, S.; Nishigawa, H.; Wei, W.; Oshima, K.; Miyata, S.I.; Ugaki, M.; Hibi, T.; Namba, S. ‘Candidatus Phytoplasma ziziphi’, a novel phytoplasma taxon associated with jujube witches’-broom disease. Int. J. Syst. Evol. Microbiol. 2003, 53, 1037–1041.
- Malembic-Maher, S.; Salar, P.; Filippin, L.; Carle, P.; Angelini, E.; Foissac, X. Genetic diversity of European phytoplasmas of the 16SrV taxonomic group and proposal of ‘Candidatus Phytoplasma rubi’. Int. J. Syst. Evol. Microbiol. 2011, 61, 2129–2134.
- Win, N.K.K.; Lee, S.Y.; Bertaccini, A.; Namba, S.; Jung, H.Y. ‘Candidatus Phytoplasma balanitae’ associated with witches’ broom disease of Balanites triflora. Int. J. Syst. Evol. Microbiol. 2013, 63 Pt 2, 636–640.
- Hiruki, C.; Wang, K. Clover proliferation phytoplasma: ‘Candidatus Phytoplasma trifolii’. Int. J. Syst. Evol. Microbiol. 2004, 54, 1349–1353.
- Davis, R.E.; Zhao, Y.; Dally, E.L.; Jomantiene, R.; Lee, M.; Wei, W.; Kitajima, E.W. ‘Candidatus Phytoplasma sudamericanum’, a novel taxon, and strain PassWB-Br4, a new subgroup 16SrIII-V phytoplasma, from diseased passion fruit (Passiflora edulis f. flavicarpa Deg.). Int. J. Syst. Evol. Microbiol. 2012, 62 Pt 4, 984–989.
- Griffiths, H.M.; Sinclair, W.A.; Smart, C.D.; Davis, R.E. The phytoplasma associated with ash yellows and lilac witches’-broom: ‘Candidatus Phytoplasma fraxini’. Int. J. Syst. Evol. Microbiol. 1999, 49, 1605–1614.
- Davis, R.E.; Zhao, Y.; Wei, W.; Dally, E.L.; Lee, M. ‘Candidatus Phytoplasma luffae’, a novel taxon associated with witches’ broom disease of loofah, Luffa aegyptica Mill. Int. J. Syst. Evol. Microbiol. 1999, 67, 3127–3133.
- Verdin, E.; Salar, P.; Danet, J.L.; Choueiri, E.; Jreijiri, F.; El Zammar, S.; Gelie, B.; Bove, J.M.; Garnier, M. ‘Candidatus Phytoplasma phoenicium’sp. nov., a novel phytoplasma associated with an emerging lethal disease of almond trees in Lebanon and Iran. Int. J. Syst. Evol. Microbiol. 2003, 53, 833–838.
- Seemüller, E.; Schneider, B. ‘Candidatus Phytoplasma mali’, ‘Candidatus Phytoplasma pyri’and ‘Candidatus Phytoplasma prunorum’, the causal agents of apple proliferation, pear decline and European stone fruit yellows, respectively. Int. J. Syst. Evol. Microbiol. 2004, 54, 1217–1226.
- Marcone, C.; Gibb, K.S.; Streten, C.; Schneider, B. ‘Candidatus Phytoplasma spartii’, ‘Candidatus Phytoplasma rhamni’ and ‘Candidatus Phytoplasma allocasuarinae’, respectively associated with spartium witches’-broom, buckthorn witches’-broom and allocasuarina yellows diseases. Int. J. Syst. Evol. Microbiol. 2004, 54, 1025–1029.
- Jung, H.Y.; Sawayanagi, T.; Wongkaew, P.; Kakizawa, S.; Nishigawa, H.; Wei, W.; Oshima, K.; Miyata, S.I.; Ugaki, M.; Hibi, T.; et al. ‘Candidatus Phytoplasma oryzae’, a novel phytoplasma taxon associated with wheat yellow dwarf disease. Int. J. Syst. Evol. Microbiol. 2003, 53, 1925–1929.
- Šafárŏvá, D.; Zemanek, T.; Valova, P.; Navratil, M. ‘Candidatus Phytoplasma cirsii’, a novel taxon from creeping thistle . Int. J. Syst. Evol. Microbiol. 2016, 66, 1745–1753.
- Davis, R.E.; Dally, E.L.; Gundersen, D.E.; Lee, I.-M.; Habili, N. “Candidatus Phytoplasma australiense,” a new phytoplasma taxon associated with Australian grapevine yellows. Int. J. Syst. Bacteriol. 1997, 47, 262–269.
- Sawayanagi, T.; Horikoshi, N.; Kanehira, T.; Shinohara, M.; Bertaccini, A.; Cousin, M.T.; Hiruki, C.; Namba, S. ‘Candidatus Phytoplasma japonicum’, a new phytoplasma taxon associated with Japanese Hydrangea phyllody. Int. J. Syst. Evol. Microbiol. 1999, 49, 1275–1285.
- Valiunas, D.; Staniulis, J.; Davis, R.E. ‘Candidatus Phytoplasma fragariae’, a novel phytoplasma taxon discovered in yellows diseased strawberry, Fragaria× ananassa. Int. J. Syst. Evol. Microbiol. 1999, 56, 277–281.
- Quaglino, F.; Zhao, Y.; Casati, P.; Bulgari, D.; Bianco, P.A.; Wei, W.; Davis, R.E. ‘Candidatus Phytoplasma solani’, a novel taxon associated with stolbur-and bois noir-related diseases of plants. Int. J. Syst. Evol. Microbiol. 2013, 63 Pt 8, 2879–2894.
- Martini, M.; Marcone, C.; Mitrović, J.; Maixner, M.; Delić, D.; Myrta, A.; Ermacora, P.; Bertaccini, A.; Duduk, B. ‘Candidatus Phytoplasma convolvuli’, a new phytoplasma taxon associated with bindweed yellows in four European countries. Int. J. Syst. Evol. Microbiol. 2012, 62 Pt 12, 2910–2915.
- Davis, R.E.; Harrison, N.A.; Zhao, Y.; Wei, W.; Dally, E.L. ‘Candidatus Phytoplasma hispanicum’, a novel taxon associated with Mexican periwinkle virescence disease of Catharanthus roseus. Int. J. Syst. Evol. Microbiol. 2016, 66, 3463–3467.
- Fernández, F.D.; Galdeano, E.; Kornowski, M.V.; Arneodo, J.D.; Conci, L.R. Description of ‘Candidatus Phytoplasma meliae’, a phytoplasma associated with Chinaberry (Melia azedarach L.) yellowing in South America. Int. J. Syst. Evol. Microbiol. 2016, 66, 5244–5251.
- Marcone, C.; Schneider, B.; Seemüller, E. ‘Candidatus Phytoplasma cynodontis’, the phytoplasma associated with Bermuda grass white leaf disease. Int. J. Syst. Evol. Microbiol. 2004, 54, 1077–1082.
- Montano, H.G.; Davis, R.E.; Dally, E.L.; Hogenhout, S.; Pimentel, J.P.; Brioso, P.S. ‘Candidatus Phytoplasma brasiliense’, a new phytoplasma taxon associated with hibiscus witches’ broom disease. Int. J. Syst. Evol. Microbiol. 2001, 51, 1109–1118.
- Arocha, Y.; Lopez, M.; Pinol, B.; Fernandez, M.; Picornell, B.; Almeida, R.; Palenzuela, I.; Wilson, M.R.; Jones, P. ‘Candidatus Phytoplasma graminis’ and ‘Candidatus Phytoplasma caricae’, two novel phytoplasmas associated with diseases of sugarcane, weeds and papaya in Cuba. Int. J. Syst. Evol. Microbiol. 2005, 55, 2451–2463.
- Lee, M.; Bottner, K.D.; Secor, G.; Rivera-Varas, V. ‘Candidatus Phytoplasma americanum’, a phytoplasma associated with a potato purple top wilt disease complex. Int. J. Syst. Evol. Microbiol. 2005, 56, 1593–1597.
- Jung, H.Y.; Sawayanagi, T.; Kakizawa, S.; Nishigawa, H.; Miyata, S.I.; Oshima, K.; Ugaki, M.; Lee, J.T.; Hibi, T.; Namba, S. ‘Candidatus Phytoplasma castaneae’, a novel phytoplasma taxon associated with chestnut witches’ broom disease. Int. J. Syst. Evol. Microbiol. 2002, 52, 1543–1549.
- Schneider, B.; Torres, E.; Martín, M.P.; Schröder, M.; Behnke, H.D.; Seemüller, E. ‘Candidatus Phytoplasma pini’, a novel taxon from Pinus silvestris and Pinus halepensis. Int. J. Syst. Evol. Microbiol. 2005, 55, 303–307.
- Harrison, N.A.; Davis, R.E.; Oropeza, C.; Helmick, E.E.; Narvaez, M.; Eden-Green, S.; Dollet, M.; Dickinson, M. ‘Candidatus Phytoplasma palmicola’, associated with a lethal yellowing-type disease of coconut (Cocos nucifera L.) in Mozambique. Int. J. Syst. Evol. Microbiol. 2014, 64 Pt 6, 1890–1899.
- Al-Saady, N.A.; Khan, A.J.; Calari, A.; Al-Subhi, A.M.; Bertaccini, A. ‘Candidatus Phytoplasma omanense’, associated with witches’-broom of Cassia italica (Mill.) Spreng. in Oman. Int. J. Syst. Evol. Microbiol. 2008, 58, 461–466.
- Zhao, Y.; Sun, Q.; Wei, W.; Davis, R.E.; Wu, W.; Liu, Q. ‘Candidatus Phytoplasma tamaricis’, a novel taxon discovered in witches’-broom-diseased salt cedar (Tamarix chinensis Lour.). Int. J. Syst. Evol. Microbiol. 2009, 59, 2496–2504.
- Lee, I.M.; Bottner-Parker, K.D.; Zhao, Y.; Villalobos, W.; Moreira, L. ‘Candidatus Phytoplasma costaricanum’, a novel phytoplasma associated with an emerging disease in soybean (Glycine max). Int. J. Syst. Evol. Microbiol. 2011, 61, 2822–2826.
- Nejat, N.; Vadamalai, G.; Davis, R.E.; Harrison, N.A.; Sijam, K.; Dickinson, M.; Abdullah, S.N.A.; Zhao, Y. ‘Candidatus Phytoplasma malaysianum’, a novel taxon associated with virescence and phyllody of Madagascar periwinkle (Catharanthus roseus). Int. J. Syst. Evol. Microbiol. 2013, 63 Pt 2, 540–548.
- Naderali, N.; Nejat, N.; Vadamalai, G.; Davis, R.E.; Wei, W.; Harrison, N.A.; Kong, L.; Kadir, J.; Tan, Y.H.; Zhao, Y. ‘Candidatus Phytoplasma wodyetiae’, a new taxon associated with yellow decline disease of foxtail palm (Wodyetia bifurcata) in Malaysia. Int. J. Syst. Evol. Microbiol. 2017, 67, 3765–3772.
- Jardim, B.R.; Kinoti, W.M.; Tran-Nguyen, L.T.; Gambley, C.; Rodoni, B.; Constable, F.E. ‘Candidatus Phytoplasma stylosanthis’, a novel taxon with a diverse host range in Australia, characterised using multilocus sequence analysis of 16S rRNA, secA, tuf, and rp genes. Int. J. Syst. Evol. Microbiol. 2021, 71, ijsem004589.
- Miyazaki, A.; Shigaki, T.; Koinuma, H.; Iwabuchi, N.; Rauka, G.B.; Kembu, A.; Saul, J.; Watanabe, K.; Nijo, T.; Maejima, K.; et al. ‘Candidatus Phytoplasma noviguineense’, a novel taxon associated with Bogia coconut syndrome and banana wilt disease on the island of New Guinea. Int. J. Syst. Evol. Microbiol. 2018, 68, 170–175.
- Jones, L.M.; Pease, B.; Perkins, S.L.; Constable, F.E.; Kinoti, W.M.; Warmington, D.; Allgood, B.; Powell, S.; Taylor, P.; Pearce, C.; et al. ‘Candidatus Phytoplasma dypsidis’, a novel taxon associated with a lethal wilt disease of palms in Australia. Int. J. Syst. Evol. Microbiol. 2021, 71, 004818.
- Zhao, Y.; Wei, W.; Davis, R.E.; Lee, M.; Bottner-Parker, K.D. The agent associated with blue dwarf disease in wheat represents a new phytoplasma taxon, ‘Candidatus Phytoplasma tritici’. Int. J. Syst. Evol. Microbiol. 2021, 71, 004604.
- Zreik, L.; Carle, P.; Bove, J.M.; Garnier, M. Characterization of the mycoplasmalike organism associated with witches’-broom disease of lime and proposition of a “Candidatus” taxon for the organism, “Candidatus Phytoplasma aurantifolia”. Int. J. Syst. Bacteriol. 1995, 45, 449–453.
- Bertaccini, A.; Arocha-Rosete, Y.; Contaldo, N.; Duduk, B.; Fiore, N.; Montano, H.G.; Kube, M.; Kuo, C.H.; Martini, M.; Oshima, K.; et al. Revision of the ‘Candidatus Phytoplasma’species description guidelines. Int. J. Syst. Evol. Microbiol. 2022, 72, 005353.
- Kirdat, K.; Tiwarekar, B.; Thorat, V.; Sathe, S.; Shouche, Y.; Yadav, A. ‘Candidatus Phytoplasma sacchari’, a novel taxon-associated with Sugarcane Grassy Shoot (SCGS) disease. Int. J. Syst. Evol. Microbiol. 2021, 71, 004591.
- Zhao, Y.; Davis, R.E. Criteria for phytoplasma 16Sr group/subgroup delineation and the need of a platform for proper registration of new groups and subgroups. Int. J. Syst. Evol. Microbiol. 2021, 66, 2121–2123.
- MacLean, A.M.; Sugio, A.; Makarova, O.V.; Findlay, K.C.; Grieve, V.M.; Tóth, R.; Nicolaisen, M.; Hogenhout, S.A. Phytoplasma effector SAP54 induces indeterminate leaf-like flower development in Arabidopsis plants. Plant Physiol. 2011, 157, 831–841.
- Wei, W.; Davis, R.E.; Nuss, D.L.; Zhao, Y. Phytoplasmal infection derails genetically preprogrammed meristem fate and alters plant architecture. Proc. Natl. Acad. Sci. USA 2013, 110, 19149–19154.
- Wu, W.; Cai, H.; Wei, W.; Davis, R.E.; Lee, I.M.; Chen, H.; Zhao, Y. Identification of two new phylogenetically distant phytoplasmas from S enna surattensis plants exhibiting stem fasciation and shoot proliferation symptoms. Ann. Appl. Biol. 2012, 160, 25–34.
- Zwolińska, A.; Krawczyk, K.; Borodynko-Filas, N.; Pospieszny, H. Non-crop sources of Rapeseed Phyllody phytoplasma (‘Candidatus Phytoplasma asteris’: 16SrI-B and 16SrI-(B/L)L), and closely related strains. Crop Prot. 2019, 119, 59–68.
- Lee, I.M.; Hammond, R.W.; Davis, R.E.; Gundersen, D.E. Universal amplification and analysis of pathogen 16S rDNA for classification and identification of mycoplasmalike organisms. Phytopathology 1993, 83, 834–842.
- Gundersen, D.E.; Lee, I.-M. Ultrasensitive detection of phytoplasmas by nested-PCR assays using two universal primer pairs. Phytopathol. Mediterr. 1996, 35, 114–151.
- Christensen, N.M.; Nicolaisen, M.; Hansen, M.; Schulz, A. Distribution of phytoplasmas in infected plants as revealed by real-time PCR and bioimaging. Mol. Plant-Microbe Interact. 1993, 17, 1175–1184.
- Wei, W.; Kakizawa, S.; Suzuki, S.; Jung, H.Y.; Nishigawa, H.; Miyata, S.; Oshima, K.; Ugaki, M.; Hibi, T.; Namba, S. In planta dynamic analysis of onion yellows phytoplasma using localized inoculation by insect transmission. Phytopathology 2004, 94, 244–250.
- Mehle, N.; Dreo, T.; Ravnikar, M. Quantitative analysis of “flavescence doreé” phytoplasma with droplet digital PCR. Phytopathogenic Mollicutes 2014, 4, 9–15.
- Dickinson, M. Loop-mediated isothermal amplification (LAMP) for detection of phytoplasmas in the field. In Plant Pathology; Humana Press: New York, NY, USA, 2015; pp. 99–111.
- Wheatley, M.S.; Wang, Q.; Wei, W.; Bottner-Parker, K.D.; Zhao, Y.; Yang, Y. Cas12a-based diagnostics for potato purple top disease complex associated with infection by ‘Candidatus Phytoplasma trifolii’-related strains. Plant Dis. 2022, PDIS09212119RE.
- Deng, S.J.; Hiruki, C. Amplification of 16S ribosomal-RNA genes from culturable and nonculturable Mollicutes. J. Microbiol. Methods 1991, 14, 53–61.
- Schneider, B.; Seemüller, E.; Smart, C.D.; Kirkpatrick, B.C. Phylogenic classification of plant pathogenic mycoplasmalike organisms or phytoplasmas. In Molecular and Diagnostic Procedures in Mycoplasmology; Razin, I.R., Tully, J.G., Eds.; Academic Press: San Diego, CA, USA, 1995; pp. 369–380.
- Casati, P.; Quaglino, F.; Stern, A.R.; Tedeschi, R.; Alma, A.; Bianco, P.A. Multiple gene analyses reveal extensive genetic diversity among ‘Candidatus Phytoplasma mali’populations. Ann. Appl. Biol. 2011, 158, 257–266.
- Wei, W.; Cai, H.; Jiang, Y.; Lee, I.M.; Davis, R.E.; Ding, Y.; Yuan, E.; Chen, H.; Zhao, Y. A new phytoplasma associated with little leaf disease in azalea: Multilocus sequence characterization reveals a distinct lineage within the aster yellows phytoplasma group. Ann. Appl. Biol. 2011, 158, 318–330.
