Interleukin-11 (IL11), a stromal-cell derived pleiotropic cytokine with profibrotic and cellular remodeling properties, as a potential biomarker in NSCLC. IL11 is an important tumor-promoting cytokine that that has both diagnostic and prognostic value in patients with NSCLC. Multiple in vitro studies confirm that IL11 activates known tumor-promoting signaling pathways and clinical studies link increased IL11 expression to poorer prognosis.
1. Interleukin-11: Member of the IL6 Family of Cytokines
Cytokines are small soluble secreted proteins that participate in autocrine, paracrine, and endocrine signaling to facilitate a diverse range of physiological functions including regulation of immunity, inflammation, cellular proliferation and cellular growth. They are often classified based on commonalities in structure, function or receptors. IL11 is classified in the IL6 family of cytokines that share commonality in the glycoprotein 130 (gp130) signaling receptor subunit, in combination with their cytokine-specific cognate receptors [
9,
10]. This family of cytokines and their cognate receptors have been regarded as important contributors to cancer biology [
11,
12,
13,
14], and may serve as potential biomarkers in disease progression. Similarly, other members of the IL6 family cytokines are known to be implicated in NSCLC as well (
Table 1).
Table 1. Other IL6 family cytokines and components of the specific receptors associated with NSCLC.
| Cytokine |
Receptors |
References |
| IL6 |
IL6R, gp130/IL6ST |
[15,16,17,18,19,20,21,22] |
| IL-31 |
IL31Rα, OSMR |
[23] |
| LIF |
LIFR/LIFRα, gp130/IL6ST |
[24] |
| OSM |
OSMR/OSMRβ, gp130/IL6ST, LIFR |
[25,26] |
| CLCF1 |
CNTFR, LIFR, gp130/IL6ST |
[22,27] |
2. Interleukin 11 Drives Pulmonary Fibrosis and Inflammation
IL11 was initially implicated in several inflammatory lung diseases such as asthma and tuberculosis infection. However, recent evidence has demonstrated that IL11 is an important determinant of fibrosis and chronic inflammation in the lung (reviewed in [
28]). Pulmonary epithelial cells and fibroblasts express high levels of IL11RA and are prominent cellular sources of IL11 in response to respiratory infections. Hence, it is proposed that IL11 acts in an autocrine/paracrine fashion in response to pathogens or after tissue injury. IL11 is upregulated in pulmonary fibroblasts from patients with idiopathic pulmonary fibrosis (IPF), a form of progressive fibrosing interstitial pneumonia characterized by an excess of activated myofibroblasts, and its elevated expression in the IPF lung is associated with fibrosis and disease severity [
29]. In vitro studies on non-transformed pulmonary cells showed that IL11 triggers the proliferation and transformation of quiescent fibroblasts into collagen-producing invasive myofibroblasts [
29], and induces an EMT program in epithelial cells. Pharmacological inhibition of IL11 or fibroblast-specific blockade of IL11 signaling reduced fibroblast invasion in vitro and reversed pulmonary fibrosis and inflammation in a murine model of IPF [
29,
30].
3. Interleukin 11 Is a Tumor-Promoting Cytokine in NSCLC
IL11 was first isolated in 1990 from a primate bone marrow-derived stromal cell line and was identified to possess hematopoietic and thrombopoietic properties [
33]. Subsequently, IL11 was evaluated as a potential therapy for chemotherapy-induced thrombocytopenia among cancer patients [
34,
35]. The need to determine suitability of IL11 treatment for thrombocytopenia in cancer patients motivated early studies evaluating its effects on cancer cells.
Early studies involving lung cancer cells concluded that IL11 did not promote tumor cell growth. Soda et al. harvested tumor cells from a heterogenous group of clinical specimens and subjected the cells to recombinant human IL11 (rhIL11) [
36]. This study defined tumor growth stimulation as a >150% survival increase in tumor colony-forming units and found that 97% of the specimens across a variety of cancers tested were not stimulated by rhIL11. Saitoh et al. utilized a murine model of lung cancer and showed that rhIL11 inhibited proliferation in vitro while not affecting the anti-tumor effects of carboplatin, mitomycin and etoposide in mice [
37]. Treating Calu-1 cells, a NSCLC epithelial cell line, with IL11 seemed to reduce DNA synthesis but not significantly [
38]. Taken together, these early studies suggest that IL11 was more likely an inhibitor, rather than a stimulator, of tumor growth.
Later studies, however, identified IL11 as a tumor-promoting cytokine instead [
39,
40,
41,
42]. It is now known that rhIL11 does not activate mouse IL11 receptor (IL11RA), but competitively inhibits binding of endogenous murine IL11 instead [
43,
44]. Hence, the use of rhIL11 on murine lung cancer is not expected to recapitulate the effect of human IL11 on human lung cancer. It is likely due to the differential effects of cross- and same-species recombinant IL11 that a later study utilizing same-species rhIL11 concluded IL11 to be tumor promoting [
45], while an earlier study that used cross-species rhIL11 concluded otherwise [
37]. Zhao et al. provided direct evidence that IL11 promotes tumor growth using lentivirus-mediated IL11 overexpression and knockout in A549 and H1299 lung cancer cell lines [
46].
IL11 signaling can occur either via classical signaling or trans-signaling. During classical signaling, IL11 binds to its specific receptor IL11RA, which leads to dimerization of the signal transducing gp130 subunit and activation of downstream signaling. IL11RA is differentially expressed in NSCLC and has been targeted in a preclinical study to reduce tumor growth [
58]. In trans-signaling, soluble IL11RA are generated from mRNA splice variants or from proteolytic cleavage of the membrane-bound receptor complexing with the cytokine extracellularly [
59]. This complex then binds to any gp130, which is ubiquitously expressed on cell surfaces, without requiring the cytokine-specific receptor potentially expanding the repertoire of effector cells. IL6 trans-signaling has been reported to be important in KRAs-driven NSCLC [
60], but it is yet uncertain whether IL11 functions similarly. Downstream of cytokine-receptor binding, JAK/STAT [
39,
40,
61], PI3K/AKT, and Ras/ERK pathways can be activated resulting in cell proliferation, inhibition of pro-apoptotic proteins, activation of anti-apoptosis proteins, and angiogenesis [
62,
63]. Activation of these pathways has been proposed as a prognostic outcome predictor for NSCLC [
64,
65,
66,
67].
4. Sources of Biomarkers in NSCLC
Biomarkers are measurable characteristics of normal and pathogenic processes or exposure to interventions [
74]. Sources for molecular biomarkers in NSCLC may include peripheral blood, bronchoalveolar lavage, breath exudate, lung tissue, pleural fluid, urine, sputum and saliva. These sources differ in feasibility, reproducibility, and procedural risk profile. Peripheral blood is readily accessible and is useful for comparisons between healthy, disease and treated populations and is also suitable in longitudinal studies measuring response to therapy. However, the sensitivity, specificity and reproducibility of blood biomarkers is limited by production by sources other than tumor, volume of distribution, and biomarker metabolism and turnover. Alternatively, molecular biomarkers identified via lung tumor tissue most directly reflect true tumor biology. However, procurement of tissue samples requires invasive methods such as diagnostic biopsy samples, post-mortem collection, or from surgical resection. Specimens from surgical resection provide the greatest quantity of tumor tissue and allows adjacent histologically normal tissue to be collected concurrently for comparisons.
5. IL11 Differential Expression in NSCLC Tumor Tissue
The Cancer Genome Atlas (TCGA) database is a commonly referenced publicly accessible dataset containing RNAseq data for tumor samples from NSCLC patients, including a proportion of matched adjacent normal samples [
75]. The Genotype-Tissue Expression (GTEx) project is a publicly available resource that provides genotype and expression data for tissue from different sources, including those from non-cancerous lung [
76]. Based on the UCSC Xena platform [
77,
78], which utilizes these datasets collectively,
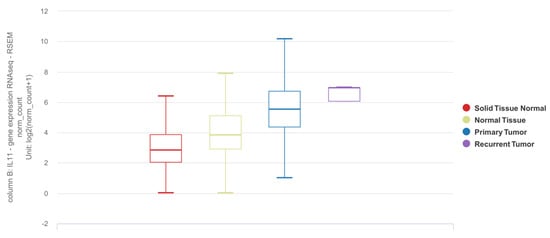
IL11 mRNA expression is elevated in tumor tissues as compared to adjacent normal or tissues from donors without lung cancer (
Figure 1).
Figure 1. Differential expression of
IL11 mRNA in lung adenocarcinoma (LUAD) and squamous cell carcinoma (SCC) based on the TCGA TARGET GTEx dataset. Solid tissue normal: adjacent normal tissue from TCGA dataset (n = 109). Normal tissue: tissue from subjects with no lung adenocarcinoma from GTEx dataset (n = 288). Primary tumor: lung tumor tissue (LUAD or SCC) from TCGA dataset (n = 1011). Recurrent tumor: lung tumor tissue from recurrence (n = 2). The results shown here are in whole or part based upon data generated by the TCGA Research Network [
75] and GTEx [
76] databases using the UCSC Xena online platform [
77,
78].
https://xenabrowser.net was accessed on 26 April 2022.
6. Detection of IL11 for Diagnosis of NSCLC
To the best of our knowledge, there are two studies that describe the utility of IL11 as a diagnostic biomarker (Table 2). Pastor et al. recruited a prospective cohort of 369 patients for which use of diagnostic biomarkers in bronchoalveolar lavage fluid (BALF) can potentially facilitate early detection of lung cancer [90]. Among 80 cytokines and growth factors, inflammation-related protein array analyses identified IL11 expression to be differentially increased in LUAD BALF samples in an initial discovery cohort, and subsequently validated in two separate exploratory and diagnostic cohorts. This study demonstrated that BALF IL11 expression was largely restricted to patients with LUAD, with or without chronic obstructive pulmonary disease (COPD). Despite the eventual diagnosis at stage III or IV in the majority of the LUAD cohort, the diagnostic performance of BALF IL11 remains similar between the subgroups even for the early stages. Interestingly, BALF IL11 expression was not increased in squamous cell carcinoma (SCC), a type of NSCLC of epithelial origin, which may suggest cell type-selective IL11 expression in NSCLC. In another study, Wu et al. measured IL11 protein concentration in serum and exhaled breath condensate in NSCLC cases compared to healthy donors and found increased IL11 expression in NSCLC patients even at early-stage disease [91].
Table 2. Studies identifying IL11 as a diagnostic biomarker.
| Study |
Recruited Population |
Comparison |
Sample Type |
Diagnostic Biomarker |
Assay |
Receiver Operator Curve and Test Metrics |
AUC
(95% CI) |
Cutoff (pg/mL) |
Sensitivity
(95% CI) |
Specificity (95% CI) |
PPV
(95% CI) |
NPV
(95% CI) |
| Pastor et al. [90] |
Age > 40 yrs, current or ex-smokers of 30 pack-years, evaluated for hemoptysis or pulmonary nodule or mass, excluding those with prior diagnosis of malignancy, active tuberculosis, history of drug abuse or other inflammatory disease apart from COPD |
LUAD vs. non-LUAD—First validation cohort (n = 149) |
BALF |
IL11 protein |
ELISA |
0.93 (0.90–0.97) |
42.0 |
90.2 (79–95.7) |
88.7 (90.6–93.5) |
80.7 (68.7–88.9) |
94.5 (87.8–97.6) |
| LUAD vs. non-LUAD—Second validation cohort (n = 160) |
BALF |
IL11 protein |
ELISA |
0.95 (0.92–0.98) |
42.0 |
90.6 (79.7–95.9) |
83.0 (86.8–87.7) |
60.8 (49.7–70.8) |
96.8 (92.7–98.6) |
| Wu et al. [91] |
NSCLC patients with no history of radiochemotherapy, immune-targeted therapy or surgery (n = 91 for serum, of which 63 have LUAD and 28 have SCC; 64 for EBC) |
Healthy volunteers without acute or chronic infectious diseases, vital organ diseases, or genetic family tumor history (n = 72 for serum; 63 for EBC) |
Serum |
IL11 protein |
ELISA |
0.93 (0.88–0.97) |
126.1 |
75.0 |
100.0 |
NR |
NR |
| EBC |
IL11 protein |
ELISA |
0.78 (0.69–0.86) |
21.5 |
78.1 |
79.4 |
NR |
NR |
This entry is adapted from the peer-reviewed paper 10.3390/cells11142257